Closer to understanding how bacteria become resistant to antibiotics
Haemophilus influenzae stands out as a significant bacterial culprit, notorious for causing severe respiratory tract infections, including pneumonia. It has remarkably developed resistance to a plethora of drugs, positioning it as a formidable challenge in medical treatment. Now researchers from Department of Biology, University of Copenhagen have made significant findings in unraveling the mechanisms behind how the resistance evolves.
Yong E Zhang from The Section of Functional Genomics, Department of Biology explains, “This antibiotic resistance of the bacteria stems from its unique ability, termed natural competence, enabling it to actively pick up external DNA from its environment and integrate it seamlessly into its own genetic makeup. This becomes particularly concerning when the external DNA includes genes that render the bacteria resistant to antibiotics”.
Grasping the workings of this process is paramount, and it’s clear that under certain conditions, the bacteria are prepared to acquire and incorporate external DNA, potentially leading to antibiotic resistance. Prior discoveries have shed light on this complex mechanism, highlighting the role of cyclic AMP (cAMP), a crucial second messenger, in kickstarting this DNA acquisition process, while purine nucleotides play a contrasting role in inhibiting it. The dynamics between cAMP and these nucleotides is intricate, yet we have made significant strides in unraveling it.
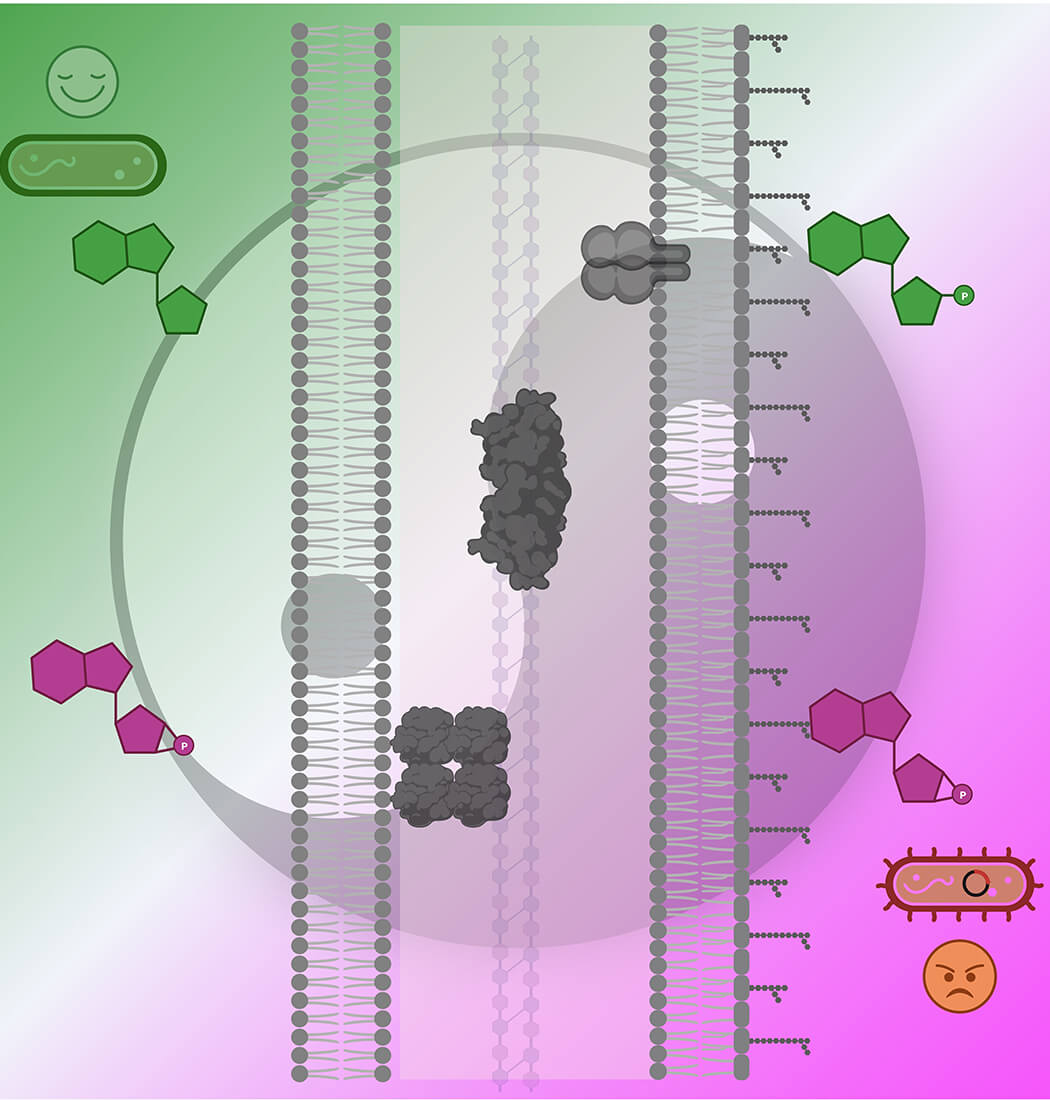
The human pathogen Haemophilus influenzae's transition between active cell growth (depicted in green) and natural competence (illustrated in magenta) are mutually regulated by the second messenger cyclic AMP (colored in magenta) and purine nucleotides (highlighted in green). This regulation occurs as they competitively bind to three periplasmic enzymes (presented as dark grey surface models).
Created with BioRender.com.
Yong E Zhang continues, “In our current study, we demonstrate that cAMP acts as a master regulator, effectively inhibiting the activity of three specific enzymes in Haemophilus influenzae. This regulatory mechanism ensures that the bacteria only initiate the natural competence state when external nucleotides are scarce, establishing a strategic link between the bacteria’s growth halt and its readiness to acquire new genetic traits”.
This discovery not only sheds light on Haemophilus influenzae’s behavior but also paves the way for understanding similar mechanisms in closely related bacterial pathogens, and even in other paramount human pathogens like Vibrio cholerae.
Link to the article in Journal of Biological Chemistry
Contact
PhD, Group leader Yong E Zhang
Laboratory of Bacterial Second Messengers and Antimicrobial Resistance and Tolerance (Bac-SMART)
Section of Functional Genomics, Department of Biology, University of Copenhagen
Email: yong.zhang@bio.ku.dk
Webpage: https://sites.google.com/view/zhangylab-ucph/home
Helle Blæsild
PR & Communication
Department of Biology, University of Copenhagen
Tel. +45 2875 2076
Mail: helleb@bio.ku.dk
