A new family tree revises our understanding of bird evolution
A team of researchers from the Bird 10,000 Genomes (B10K) consortium has reported a new bird family tree that promises to reshape our understanding of avian evolution. Published in Nature on April 1st, 2024, the study entitled "Complexity of avian evolution revealed by family-level genomes" marks a pivotal advancement in the long-contested evolutionary relationships among living birds.
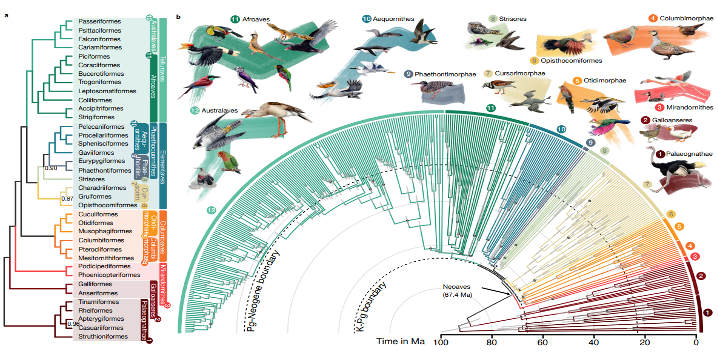
Birds are the only dinosaur lineage that survived until today. About 66 million years ago at the Cretaceous–Paleogene (K–Pg) boundary, a mass extinction event destroyed all non-avian dinosaurs, providing an opportunity for birds to diversify rapidly and occupy a wide range of ecological niches. Neoaves, a diverse group comprising approximately 95% of all bird species today, are thought to have emerged from this radiationin a ‘Big Bang’, although some research indicates an earlier origin of the group. From the towering condors of the Andes to the diminutive hummingbirds flitting through tropical forests, Neoaves encompass a stunning diversity of forms and functions. Despite considerable efforts to reconstruct avian evolutionary history and the impacts of the K-Pg event using morphological data and molecular data, the precise branching order and relationships among the neoavian lineages remained contentious.
By employing full genome data across 363 bird species, the B10K consortium produced the largest-ever dataset used for phylogenetic analyses of birds. The team built a new pipeline to extract over 150,000 regions spread out across the genome. “We characterized phylogenetic relationships across the entire genome and identified patterns associated with the genomic context and sequence characteristics”, said Josefin Stiller, the leading author of this study and an Assistant Professor in evolutionary biology at the Department of Biology, University of Copenhagen, “We found that various parts of the genome, for example individual chromosomes or protein-coding genes, often support vastly different trees. This likely explains why studies that only analyzed certain genomic parts were in conflict”.
Putting together high quality and large amounts of data was key to producing a robust phylogenetic tree. The team discovered that for most branches, a consensus on their relationships can be reached when a sufficient amount of data was used. But the phylogenetic positions for some bird groups like owls and hawks remain puzzling even with a full-genome scale of data. “More data does not necessarily produce a better solution”, said Guojie Zhang from Zhejiang University and Adjunct Professor at the University of Copenhagen . Siavash Mirarab, co-senior author of the study, a professor of electrical and computer engineering at the University of California, San Diego, added that, “The reason for this may be some complex evolutionary history like ancient cross-mating between two lineages, incomplete lineage sorting, long-branch attraction, and biased DNA sequence content, all of which can interfere with the reconstruction of phylogenetic trees.”
The study also proposes a more accurate time scale for the diversification of modern birds, lending weight to the ‘Big Bang’ scenario of a rapid radiation at or near the mass extinction at the K–Pg boundary. The researchers found that this radiation coincided with remarkable genetic and morphological changes among birds, including greater mutation rates, smaller body sizes, and larger brains, and larger effective population sizes. “This illustrates the power of comparative genomics: by comparing genomes of living species, we can uncover traces of events that happened 66 million years ago,” Stiller said.
“Our work has changed many traditional views on the evolutionary history of birds. This new family tree will serve as a solid backbone for mapping the evolutionary history of all bird species with important implications for ornithological research and biodiversity studies,” Zhang concluded.
More information:
The Bird 10,000 Genomes (B10K) Project is an initiative aiming to map the genomes of all approximately 10,500 existing bird species. This ambitious project seeks to construct an all-encompassing avian tree of life from a whole-genome perspective, decoding the links between genetic variation and trait differences, unraveling the molecular evolution, biogeography, and biodiversity interrelations, assessing the impact of environmental changes and human activities on species evolution and biodiversity, and revealing the population history of the entire avian group.
The B10K was initiated by Professor Guojie Zhang from Zhejiang University, along with professors M. Thomas P. Gilbert from University of Copenhagen, Erich D. Jarvis from The Rockefeller University, FuminLei from Chinese Academy of Sciences, Carsten Rahbek from University of Copenhagen, and Gary R. Graves from the National Museum of Natural History Smithsonian Institution. The B10K consortium brings together hundreds of experts across the globe that study various aspects of avian biology. For more information, visit the website: https://b10k.com.
The B10K Project: https://www.youtube.com/watch?v=n6skxzr7_G4
Nature article: https://www.nature.com/articles/s41586-024-07323-1
Contact
Josefin Stiller
Assistant Professor
Villum Young Investigator / Villum Centre for Biodiversity Genomics
University of Copenhagen
Mail: josefinstiller.com
Tag: @Rubyseadragon
Professor Guojie Zhang
Centre for Evolutionary & Organismal Biology, Zhejiang University
Mail: guojiezhang@zju.edu.cn
Tel: +86 159 1414 0493
Helle Blæsild
PR & Communication, Department of Biology
Mail: helleb@bio.ku.dk
Tel: +45 2875 2076
