Characterization of bacterial mercury resistance in High Arctic bacteria
Project type: Master project
Keywords
Molecular biology techniques, Next generation sequencing, Bioinformatics, Evolution (phylogeny) of genes, Global mercury cycling, Bacterial mercury resistance
 Introduction
Introduction
Mercury (Hg) is a toxic heavy metal that occurs naturally as elemental mercury (Hg0), oxidized mercury (Hg2+) and organic mercury (methyl-Hg).
Arctic food webs are becoming increasingly contaminated with mercury resulting in elevated exposure levels of indigenous human populations. It is estimated that more than 300 tons of Hg is deposited annually in the Arctic.
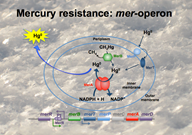
Bacteria may respond to the toxicity of Hg2+ by transforming it to gaseous Hg0 resulting in a detoxification of the immediate environment of the bacteria. The most common mechanism is the enzymatic reduction of Hg2+ to Hg0 by the mercuric reductase (MerA) encoded by the merA gene. merA is part of the mer operon that encodes a group of proteins involved in the detection, scavenging, transport and reduction of Hg. Because bacterial reduction of Hg2+ leads to the formation of volatile Hg0, mercury-resistant bacteria may act as re-emitters of Hg to the atmosphere and, hence, contribute to the detoxification of the contaminated arctic environment.
In contrast to temperate regions, little is known about the occurrence, diversity and distribution of merA genes among arctic bacterial communities.
The project Mercury resistant bacteria have previously been isolated from snow, sea ice and a freshwater lake in NE Greenland, about 900 km south of the North Pole. The bacteria were tested for their ability to transform Hg2+ to Hg0. Surprisingly, some of the highly resistant isolates appeared not to carry merA and not to produce Hg0, while others produced Hg0 but did not contain merA. The lack of detection of merA in the resistant bacteria may be due to the presence of hitherto unknown resistance mechanims or the presence of merA genes having a very low sequence similarity to known gene sequences.
Mercury resistant bacteria have previously been isolated from snow, sea ice and a freshwater lake in NE Greenland, about 900 km south of the North Pole. The bacteria were tested for their ability to transform Hg2+ to Hg0. Surprisingly, some of the highly resistant isolates appeared not to carry merA and not to produce Hg0, while others produced Hg0 but did not contain merA. The lack of detection of merA in the resistant bacteria may be due to the presence of hitherto unknown resistance mechanims or the presence of merA genes having a very low sequence similarity to known gene sequences.
The purpose of the project will be to identify and characterize the (potentially new) resistance mechanisms of selected mercury resistant bacteria from the High Arctic. The work will involve testing of the ability of the bacteria to volatilize Hg2+, detection of merA by PCR, whole-genome sequencing of the bacteria, annotation of the sequences, identification/characterization of the operons involved in the mercury resistance, and phylogenetic analyses of the genes.
 More information
More information
http://www.astrobio.net/topic/origins/extreme-life/arctic-bacteria-show-long-evolution-toxic-mercury-resistance/
Moller, A. K., et al. (2011). "Diversity and characterization of mercury-resistant bacteria in snow, freshwater and sea-ice brine from the High Arctic." Fems Microbiology Ecology 75(3): 390-401.
Moller, A. K., et al. (2014). "Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine." Fems Microbiology Ecology 87(1): 52-63.
Supervisors
Niels Kroer (nk@bio.ku.dk)
Søren J. Sørensen (sjs@bio.ku.dk)
Section of Microbiology

Contact
Section of Microbiology
Universitetsparken 15, build. 1, 1. floor
DK-2100 Copenhagen
SUPERVISORS
Professor Niels Kroer
E-mail: nk@bio.ku.dk
