CRISPRs on Plasmids: Fight Fire with Fire
Project type: Masters
Main area: CRISPR; Plasmids; Microbiology; Bioinformatics
Project Description:
CRISPR/Cas systems are prokaryotic adaptive immune systems which target foreign genetic elements such as viruses and plasmids. The application of these systems is revolutionizing gene editing both in research but also in the clinical setting. Given the novelty and great diversity within these systems, still very little is known about their mechanism and evolutionary role in natural environments. Although much attention has been given to their importance in combating phage predation, their impact on the interplay between host and plasmids is generally overlooked. Plasmids play an important role in bacterias ability to quickly acquire new abilities (ie. antibiotic resistance), and one could imagine that a trade-off between an effective immune-system and the ability to uptake beneficial plasmid-encoded traits is a defining feature of pathogenic vs. benign bacteria. However, the playing field is even more complex, as some plasmids are known to encode CRISPR/Cas system components, whilst others may potentially encode antiCRISPR proteins.
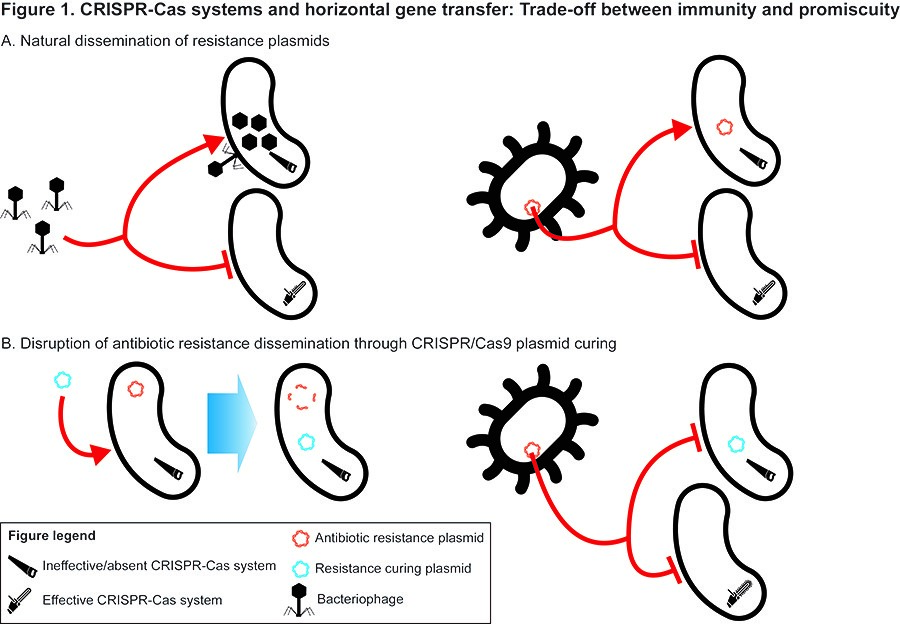
The goal of this project is to bioinformatically describe CRISPR/Cas systems on plasmids and using selected plasmids with defining CRISPR/Cas related features, experimentally investigate the interplay between promiscuity and plasmid uptake. Another key part will be investigating novel CRISPR/Cas system found on the plasmids experimentally.
You will be working with a team containing both bioinformaticians and experimentalists.
Methods used:
You will have a great deal of influence of the direction of your project, but it can include Bioinformatics, Data Analysis and Systems Biology, Molecular Microbiology, Flow Cytometry
Supervisors:
Jonas Stenløkke (jsmadsen@bio.ku.dk),
Joseph Nesme (joseph.nesme@bio.ku.dk),
Rafa Pinilla Redondo (rafael.pinilla@bio.ku.dk)
Section of Microbiology

Contact
Section of Microbiology
Universitetsparken 15, build. 1, 1. floor
DK-2100 Copenhagen
SUPERVISORS
Tenure track assistant prof. Jonas Stenløkke Madsen
E-mail: jsmadsen@bio.ku.dk
Postdoc Joseph Nesme
E-mail: joseph.nesme@bio.ku.dk
Postdoc Rafael Pinilla
E-mail: rafael.pinilla@bio.ku.dk
