Early life microbiome and later disease development
Project type: Masters
Background:
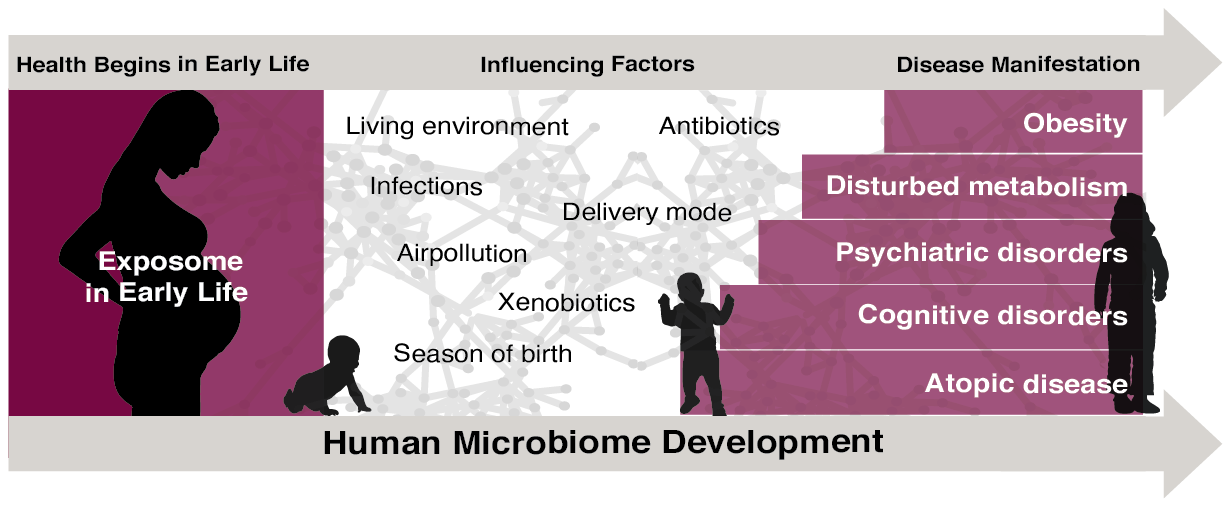
Asthma, eczema, and allergy (atopic diseases) are the first common chronic diseases to manifest during childhood. Approximately 20% of children develop asthma-like symptoms, and an estimated 300 million people suffer from asthma worldwide. A healthy embryo is considered essentially sterile and the first and very important colonization is established by its early contact with the environment. Therefore, the first years of our lives represent a critical period for susceptibility to environmental exposures – an open window – from which lasting effects can be imprinted on the developing immune system, which may in time lead to atopic disease. In collaboration with Copenhagen Prospective Studies on Asthma in Childhood (COPSAC), we study how the microbiome is an intermediary player in the interaction between the host and its environment, with a key focus in research into the extrinsic mechanisms that determine the transition from health to chronic disease.
COPSAC2010 is an ongoing Danish cohort study of 738 unselected pregnant women and their 700 children followed from pregnancy week 24. The foundation of the project will be the vast amount of data from the COPSAC clinical birth cohort with extensive longitudinal microbial samples characterized by sequencing through the first year of life (1 week, 1 month, and 1 year), as well as later time points. The primary endpoints: asthma, eczema and allergy have been diagnosed prospectively by the COPSAC pediatricians.
Types of projects:
The applicant will join the Section of Microbiology and with the microbial expertise of Professor Søren J. Sørensen’s lab, where the project will leverage a strong interdisciplinary research cooperation to pursue projects that:
- Investigate the early life exposome, which through gene-environment interactions cause immune deregulation, inflammation, and subsequent symptomatic disease.
- Examine the correlations between the early-life gut microbiome, antimicrobial resistome and later disease
- Identify microbial interaction networks and other biological markers that are key for neurodevelopment and atopic disease.
- Develop synthetic microbial communities that can be used as part of future intervention strategies against later disease development.
Techniques involved:
- Bioinformatics (R and/or Python)
- Multivariate statistics
- Classical microbiology and molecular microbiology
- Animal experiments
- High resolution imaging (FACS and CLSM)
- DNA and RNA sequencing
For more information, please contact:
Urvish Trivedi (urvish.trivedi@bio.ku.dk)
Søren Sørensen (sjs@bio.ku.dk)
READ MORE AT: https://www1.bio.ku.dk/english/research/microbiology/
Section of Microbiology

Contact
Section of Microbiology
Universitetsparken 15, build. 1, 1. floor
DK-2100 Copenhagen
 SUPERVISOR
SUPERVISOR
Professor Søren Sørensen
E-mail: sjs@bio.ku.dk
Contact
Section of Microbiology
Universitetsparken 15, build. 1, 1. floor DK-2100 Copenhagen
Assistant Professor Urvish Trivedi
E-mail: urvish.trivedi@bio.ku.dk

