Regulation of plasmid mediated biofilm formation
Project type: Master or Bachelor
Many bacterial species are known to attach to surfaces or other cells and establish biofilms which offers them several advantages compared to the planctonic lifestyle. First, the biofilm mode may facilitate synergistic interactions and gene transfer among different species. Second, biofilms may be crucial for bacterial survival in many environments, including humans, where biofilms are involved in various persistent infections. Consequently, the formation of bacterial biofilms can cause both health and environmental related problems making biofilm research an important issue.
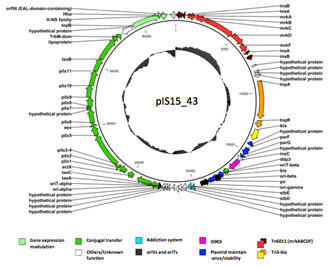
Today many plasmids occuring in the natural environment are known to be involved in biofilm formation. We have isolated a conjugative plasmid, pIS15_43, and strains carrying this plasmid have been identified as being very good biofilm formers. From subsequent annotation of the plasmid we know that it belongs to the family of IncX1 plasmids and surprisingly seems closely related to another well-known IncX1 plasmid, pOLA52, that recently was found to be involved in biofilm formation (Norman et al., 2008). Like pOLA52, the pIS15_43 plasmid carries the mrk genes encoding the type 3 fimbriae, known to be involved in biofilm formation in Klebsiella (Burmølle et al., 2008). Another interesting feature of this plasmid is the presence of pieces of the regulatory domains, EAL, HN-S and Hha, upstream the mrk genes. These domains are recognized as being indirectly involved in biofilm formation through regulation of the intracellular level of the second messenger cyclic-di-GMP (Hengge, 2009).
However, a degenerated version of EAL lacking normal enzyme activity is found on plasmid pIS15_43 and many other IncX1 plasmids. Despite being degenerated, this domain may play a central role for plasmid mediated biofilm formation. Recent experiments indicate that the function of degenerated EAL may be the opposite of the function of the intact version of EAL, as knocking out the degenerated EAL domain on plasmid pIS15_43 has resulted in a decrease in biofilm formation. The question is whether the degenerated EAL domain acts as a transcriptional regulator of the mrk gene casette? In this project molecular microbial approaches including gene cloning, gene knock-outs and complementation studies will be designed to further investigate the regulatory role of the degenerated EAL domain.
References:
Norman, A. et al. (2008). Plasmid, 60, p. 59-74.; Burmølle, M. et al. (2008). Microbiology, 154, p. 187-95 and Hengge, R. (2009). Nature Reviews, 7, p. 263-273.
Supervisors:
Jonas Stenløkke Madsen (jsmadsen@bio.ku.dk)
Section of Microbiology

Contact
Section of Microbiology
Universitetsparken 15, build. 1, 1. floor
DK-2100 Copenhagen
SUPERVISOR
Tenure track assistant prof. Jonas Stenløkke Madsen
E-mail: jsmadsen@bio.ku.dk
