CoDoN - Copenhagen DNA Analysis Center
CoDoN is an infrastructure grant, and it will provide access to new discoveries, research methodologies and applications derived from long-read sequencing to biomedical, clinical, biotechnological, natural science research & development in the Copenhagen area. It will ensure that non-specialists can explore these new research avenues without advanced laboratory or analytical infrastructure.
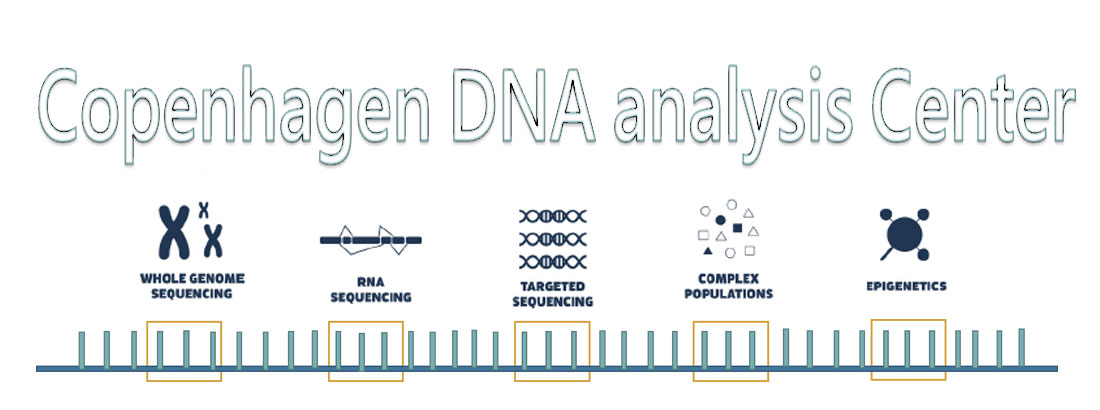
The Copenhagen DNA Analysis Center will include all necessary equipment and software to cover all procedures from sample preparation, sequencing, bioinformatic analysis and secure storage of results. The physical components include: A Bluepippin for high molecular weight DNA fragment size selection and purity assurance, a Qubit Quantitation platform for sample quantification, a Nanodrop 8000 spectrophotometer, a PacBio Sequel II for LRS, a UPS and local computers.
PacBio SMRT Sequencing produces long continuous reads that can cover large complex repetitive regions to identify and resolve structural breakpoints. It provides better accuracy using inter & intramolecular consensus sequencing. The latter uses the ‘circular consensus’ method sequencing a single molecule a number of times. This is critical for complex population applications such as resolving microbial communities, viral populations, immune repertoires, and somatic mutations where the accuracy of de novo assembly is crucial in the absence of a reference sequence. PacBio highly accurate HiFi reads can achieve Sanger-quality sequencing free of systematic error and achieve 99.999% consensus accuracy. Because DNA fragments are sequenced as single molecule without prior amplification, bias based on GC content is reduced and uniform sequencing coverage of high-/low- GC regions is enabled.
SMRT Sequencing has the ability to detect modifications within the native DNA strand enabling epigenetic modification detection at the single nucleotide and genomic region levels. Such methods can examine eukaryote epigenetics for genomic approaches to cancer and for determination in developmental biology and biomedicine. The ability to detect epigenetic modification without bias simultaneously than nucleotide sequencing eliminates the need for special sample preparation or additional sequencing reducing costs for several users.
PacBio has the highest sensitivity (>95%) for the >20,000 SVs in a human genome and the lowest false discovery rate (<5%), reducing time and cost for validation. For microbial sequencing
‘multiplexing’ microbial assay and analysis allows for the sequencing of multiple microbial isolates per Sequel II SMRT cell 8M, reducing cost per isolate. The PacBio Sequel II system allows sequencing of up to 2 million very high accuracy reads per run, which allows for example the simultaneous sequencing of up to 48 microbial genomes in a single run.
For more information please contact Assistant profesoor Joseph Nesme, E-mail: joseph.nesme@bio.ku.dk
We have dedicated specialists whom will advise on initial experimental strategies and research parameters, follow the procedure and to explain the results to make the sequencing run as efficient as possible. The facility will provide analysed results relating to the research questions and hypothesis defined instead of difficult to interpret raw sequencing data. The Copenhagen DNA Analysis Center will also assist on cross disciplinary application and accessibility.
So if your are interested in exploring your possiblities, you are very wolcome to contact Assistant professor and Coordinator Joseph Nesme, E-mail: joseph.nesme@bio.ku.dk
Professor and head for Section of Microbiology
Søren Sørensen
Universistetsparken 15, build. 1., 1st floor
dK-2100 Copenhagen
E-mail: sjs@bio.ku.dk
 Assistant professor Joseph Nesme
Assistant professor Joseph Nesme
Section of Microbiology
Universitetsparken 15, building 1, 1st floor
DK-2100 Copenhagen
E-mail: joseph.nesme@bio.ku.dk
![]() Professor and head Genome Research and Molecular Biomedicine
Professor and head Genome Research and Molecular Biomedicine
Karsten Kristiansen
Universitetsparken 13
DK-2100 Copenagen
E-mail: kk@bio.ku.dk
 Professor Guojie Zhang
Professor Guojie Zhang
Center for Ecology and Evolution
Universitetsparken 3
DK-2100 Copenhagen
E-mail: Guojie.Zhang@bio.ku.dk
![]() Professor Tom Gilbert
Professor Tom Gilbert
Section for Evolutionary Genomics
Øster Farimagsgade 5, 7, Building: 7.203
DK-1353 København K
E-mail: tgilbert@sund.ku.dk
![]() Associate professor Gisle Vestergaard
Associate professor Gisle Vestergaard
Department of Health Technology Bioinformatics, DTU
Kemitorvet, 204, 251
DK-2800 Kgs. Lyngby
E-mail: gisves@dtu.dk
 Associate professor Mani mozhiyan Arumugam
Associate professor Mani mozhiyan Arumugam
NNF Center for Basic Metabolic Research
Blegdamsvej 3B, Maersk Tower 8. s,
DK- 2200 Copenhagen
E-Mail: arumugam@sund.ku.dk
Section of Microbiology

Contact
 Professor Søren Sørensen
Professor Søren Sørensen
Universitetsparken 15,
Building 1, 1. floor
DK-2100 Copenhagen
E-mail: sjs@bio.ku.dk
 Assistant professor Joseph Nesme
Assistant professor Joseph Nesme
Universitetsparken 15,
Building 1, 1. floor
DK-2100 Copenhagen
E-mail: joseph.nesme@bio.ku.dk

