Gene expression in multispecies biofilms - linking structure and function
In this project, we will study biofilms composed of multiple species; specifically aiming at identifying the matrix components that are differently expressed in mono vs. multispecies biofilms.
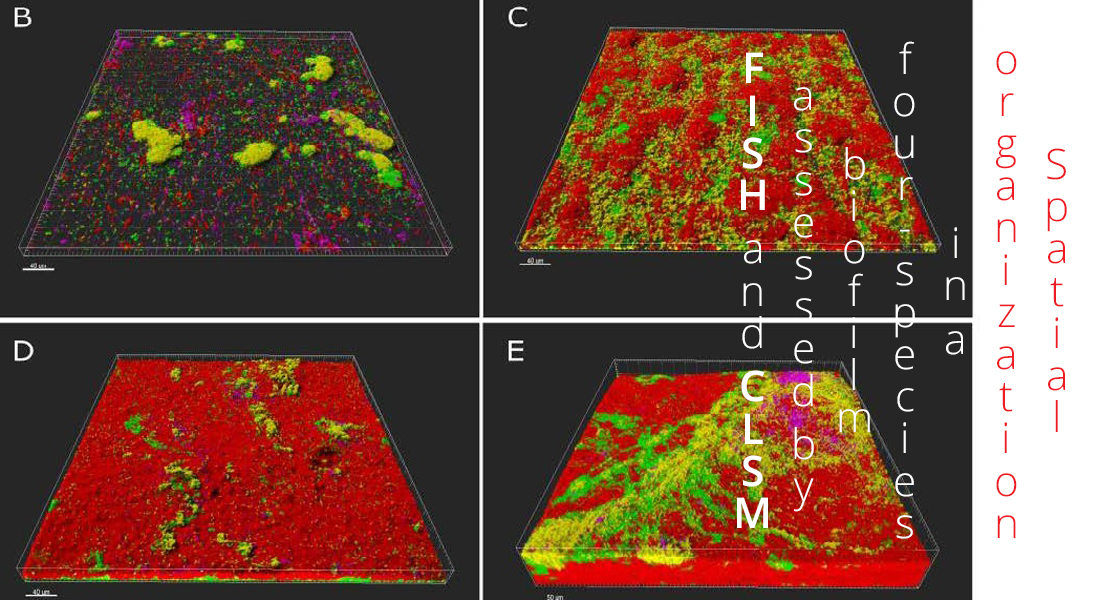
This project will advance the understanding of multispecies biofilm at the micro-scale. We will analyze the expression of matrix encoding genes in multispecies biofilms and link this to the spatial organization of the individual members.
Gene expression in multispecies biofilms - linking structure and function
Multispecies biofilms are ubiquitous in natural and man-made environments. The development and activity of these bacterial communities are shaped by interactions between residing organisms, which give rise to unique properties, such as enhanced tolerance and productivity; Multispecies biofilms are therefore both industrially challenging and biotechnologically relevant. Because of their ubiquity and high persistence, biofilms challenge various human activities. Consequently, intensive research efforts have been directed towards the biofilm area, but efficient anti-biofilm strategies are still missing. This is likely caused by a biased focus in biofilm research towards simple biofilms, composed of only one species, that neglect important interactions and synergies occurring between bacteria of different species: Only by studying the bacteria when living with other species in close proximity, will we be able to understand these communities and their underlying interactions.
In this project, we will link gene expression to biofilm function and structure, which will improve understanding of these complex communities and contribute to the development of improved eradication and application strategies.
Gains and perspectives
This project will advance the understanding of multispecies biofilm at the micro-scale. We will analyze the expression of matrix encoding genes in multispecies biofilms and link this to the spatial organization of the individual members.
We will thus be able to describe the bacterial interactions and how these interactions shape the expression of specific genes, both in relation to position in the biofilm and the other species present - all at the micro-scale level. Specifically, matrix components important for biofilm synergy and emergent properties will be identified. This is timely and highly relevant, as multispecies biofilm research is now at the stage, where the impact and unique characteristics of these complex communities are recognized, but the underlying molecular interactions are unknown.
The novel knowledge generated will serve as a platform for further research and for biotechnological applications; Multispecies biofilms are currently applied in waste water treatment, generation of biogas, bioremediation etc. and the potential for further exploration of these bacterial consortia in the transition towards a new “biobased society” is enormous. In addition, the findings og this research will likely identify regulators of emergent properties which are potential targets for development of novel anti-biofilm strategies applied in various industries challenged by biofilms, including food production facilities, pipelines and shipping, as costs and fuel consumption are enhanced due to biofilm formation on ship vessels, so efficient anti-biofilm strategies will benefit the industry and society economically as well as environmentally.
 Associate professor Mette Burmølle
Associate professor Mette Burmølle
My main research interest is the interplay and interactions among bacteria of different species within diverse communities. It is becoming more and more clear that the activity of bacteria are highly dependent on and determined by the microbial community they live in, and that the total functions of a multispecies bacterial community can’t be explained by examining each part of the community in isolation. Thus, a deeper exploration of the interactions shaping these communities is vital for understanding bacterial activity, physiology, function and evolution.
My current research aims at understanding the prevalence and underlying mechanism of interspecific interactions; both with respect to interaction characterization (synergism vs. antagonism), main facilitator (co-metabolism, co-aggregation etc.) and molecular mechanism. This includes characterisation of the impact of quorum sensing and horizontal gene transfer. I study bacterial interactions in strain collections from various natural and human health-related environments including soil, marine, freshwater, day care facilities and food processing environments.
In most cases, tight bacterial associations in a structured community are prerequisites for the interactions to develop; therefore biofilms are perfect settings for studies of interspecific bacterial interactions. I my group, different types of biofilm models, including static, batch biofilms and the newly established biofilm flow system, Bioflux 1000z, have become our model systems of choice for study of multispecies interactions.
Section of Microbiology

Funded by:
Naval Research Global grant from the Office of Naval Research - Science and Technology
Project title: Gene expression in multispecies biofilms - linking structure and function
Project period: Nov. 2019 - Oct. 2022
Contact
 Associate professor
Associate professor
Mette Burmølle
Universitetsparken 15
Building 1, 1st floor
DK-2100 Copenhagen
Email: burmolle@bio.ku.dk

