Bac-SMART – The Zhang Lab
The Bac-SMART Group (Bacterial Stress Modulation and Antimicrobial Resistance/Tolerance) decodes how bacteria use stress responses to resist antibiotics and phages. We develop novel strategies and therapeutic targets to combat persistent, untreatable infections (by e.g., CRE, XDR-Shigella).
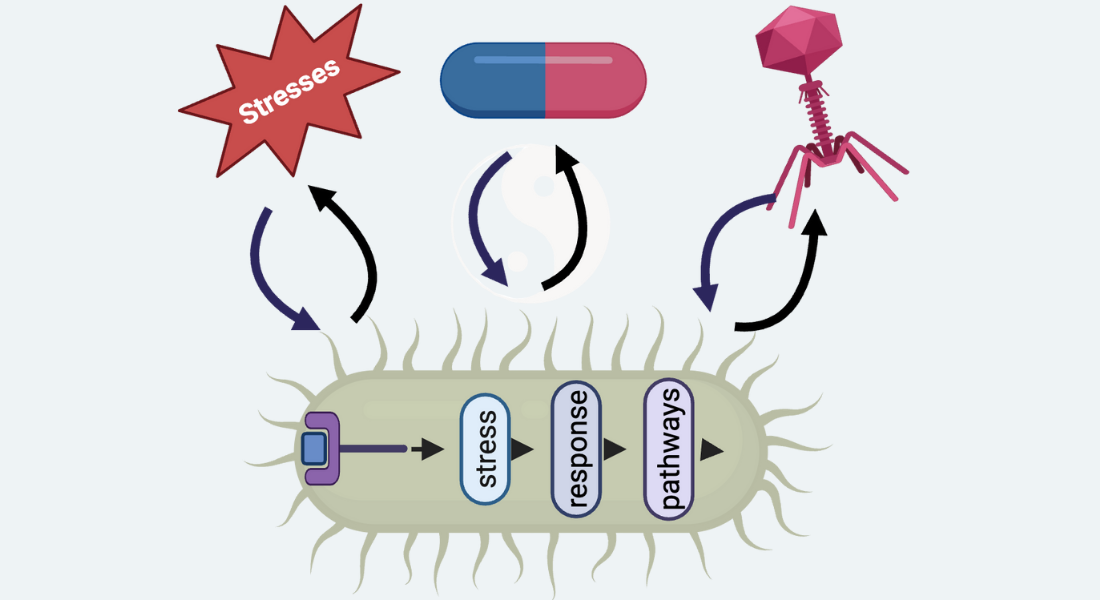
Our lab investigates how bacterial stress response pathways – governed by second messengers (e.g., (p)ppGpp, cyclic AMP) and two-component systems – enable survival under antibiotic treatment and phage predation. These pathways are fundamental drivers of antimicrobial tolerance, persistence, resistance, and the evolutionary dynamics of phage-bacteria interactions.
To study these systems, we combine molecular and biochemical techniques, systems biology, omics, structural tools (including crystallography, protein design), biophysics, and infection models (including animal studies) to dissect the mechanisms linking stress signaling to treatment failure. Through close international collaborations, we aim to reveal mechanistic insights and translate them into novel strategies to combat persistent and untreatable bacterial infections.
1. Stringent Response (SR) Pathway & Bacterial Stress Adaptation
We investigate how the universal alarmone ppGpp reprograms bacterial metabolism and physiology to survive stresses (e.g., nutrient starvation, antibiotics, virulence challenges). Key focuses include:
- ppGpp Homeostasis: Molecular regulation of RelA/SpoT enzymes that synthesize/degrade ppGpp in response to environmental cues.
- ppGpp Targets1,6,8,9: Mechanistic studies of ppGpp interactions with conserved cellular targets (e.g., PpnN-mediated metabolic balancing during stress).
- Phenotypic Outcomes: Linking ppGpp dynamics to antibiotic persistence8, resistance evolution5, and infection outcomes7.
- Single-Cell Heterogeneity5: How ppGpp-driven cell-to-cell variation enables bacterial survival under stress?
2. Phage-Bacteria Interactions & Evolutionary Arms Races2
We dissect molecular mechanisms governing phage infection under stresses, including:
- Host Hijacking: Identification of key bacterial/phage proteins enabling invasion (e.g., SR and anti-SR systems).
- Stress-Driven Coevolution: How bacterial resistance mutations (e.g., surface modification) and phage counter-adaptations shape long-term evolutionary dynamics.
- Persistent Infection Strategies: Phage exploitation of bacterial dormancy/persistence states for survival.
3. Translational Applications
Leveraging SR (,etc) and phage-host insights to develop:
- Novel Antimicrobials: Targeting ppGpp networks3 to combat multidrug-resistant infections.
- Phage-Based Solutions: Precision biocontrol agents for agriculture and engineered therapies against persistent pathogens.
- René L. Bærentsen#, Kristina Kronborg#, Ditlev E Brodersen, Yong E. Zhang*. Catalytic Mechanism and Differential Alarmone Regulation of a Conserved Stringent Nucleosidase 2025-04-14. bioRxivDOI: 1101/2025.04.13.648610
Significance: We revealed a highly conserved mechanism of nucleotide metabolism between bacterial stringent protein PpnN and plant LOG proteins producing the crucial growth factor cytokinin. -
Zhang, Y.E. Protein Kinases in Mediating Phage-Bacteria Interactions. Kinases Phosphatases 2025, 3,14. https://doi.org/10.3390/ kinasesphosphatases3030014
- Muriel Leandra Schicketanz, Magdalena Petrova, Dominik Rejman, Margherita Sosio, Stefano Donadio, Yong E. Zhang*. Direct detection of stringent alarmones (pp)pGpp using malachite green. 2024 Microbial Cell11: 312-320. doi: 15698/mic2024.08.834
Significance: This work identified and established a novel application of a well-established method, facilitating the straightforward identification of new small molecules targeting ppGpp metabolism. Given ppGpp's central role in regulating bacterial antibiotic tolerance, this approach enables large-scale screening for compounds to counter this tolerance mechanism. Three such active compounds were successfully identified. - Kristina Kronborg, Yong E. Zhang*. Cyclic AMP competitively inhibits periplasmic phosphotases to coordinate nutritional growth and competence of Haemophilus influenzae. J Bio Chem. 2023 Oct 26. DOI: 1016/j.jbc.2023.105404
(Editors’ Pick)
Significance: This work resolves a long-standing question in Haemophilus influenzae competence by identifying three novel cAMP-regulated proteins that control external DNA uptake, a key process for spreading antibiotic resistance genes. This discovery reveals a fundamental mechanism with high potential to operate similarly in major human pathogens like Vibrio cholerae. - Paulina Katarzyna Grucela, Yong E. Zhang*. Basal level of ppGpp coordinates Escherichia colicell heterogeneity and ampicillin resistance and persistence. Microbial Cell. 2023 Oct 25; 10(11): 248-260. doi: 10.15698/mic2023.11.808.
Significance: We have discovered that basal ppGpp levels, despite being extremely low, play a fundamental role in governing cell heterogeneity. This novel mechanistic insight demonstrates how heterogeneity directly influences both antibiotic resistance and bacterial persistence, challenging established assumptions about ppGpp function. - Paulina Katarzyna Grucela, Tobias Fuhrer, Uwe Sauer, Yanjie Chao* and Yong E. Zhang*. Ribose 5-phosphate: the key metabolite bridging the metabolisms of nucleotides and amino acids during stringent response in Escherichia coli? Microbial Cell. 2023 Jun 1;10(7):141-144. doi: 10.15698/mic2023.07.799. eCollection 2023 Jul 3.
Significance: We proposed a comprehensive, potentially highly conserved, model integrating the transcriptional and metabolic aspects of ppGpp coordinating rapid bacterial physiological shifts for optimized stress survivals. - Y Elizabeth Chau, Deyanira Pérez-Morales, Wael Elhenawy, Víctor H. Bustamante, Yong E. Zhang, Brian K. Coombes* (p)ppGpp-dependent regulation of the nucleotide hydrolase PpnN confers complement resistance in Salmonella entericaserovar Typhimurium. Infection & Immunity. 2021 Jan 19;89(2):e00639-20. doi: 10.1128/IAI.00639-20.
Significance: In collaboration with Dr. Brian K. Coombes we discovered a novel regulatory mechanism where ppGpp and PpnN govern Salmonella's resistance to the human complement system, revealing a previously unrecognized aspect of bacterial immune evasion. - Zhang E. Yong#*, Bærentsen L Rene#, Tobias Fuhrer, Uwe Sauer, Kenn Gerdes, Brodersen E. Ditlev*. (p)ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Molecular Cell(2019). 74(6):1239-1249.e4 (*co-corresponding author; # equal contribution))
Significance: This study uncovers a previously unknown, conserved mechanism where ppGpp and PpnN co-regulate rapid bacterial growth and antibiotic persistence development. The discovery of PpnN's essential role in this dual control reveals a fundamental aspect of bacterial complexity in stress survival. These findings provide a novel framework for understanding bacterial resilience and suggest new avenues for targeting multidrug-resistant pathogens. - Yong Zhang*, Eva Zborníková, Dominik Rejman and Kenn Gerdes*. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. 2018. mBio9:e02188-17. (*co-corresponding author)
Significance: This work achieves the first systematic identification of numerous novel proteins in E. coli that bind and metabolize ppGpp. Representing one of the most significant advance in understanding this molecule since its discovery over 50 years ago, it establishes a foundational resource for probing ppGpp's fundamental role in bacterial stress responses.
Currently (@Jun2025) open positions: one PhD, two Postdoc
We are always happy to hear from highly motivated Bachelor, Master, PhD students, and Postdoctoral researchers interested in joining our group. If you would like to be part of our team, please reach out to Yong Zhang directly at yong.zhang@bio.ku.dk. In your inquiry, kindly include a brief motivation letter, a short CV, and a description of your research interests or project ideas.
We especially encourage talented PhD students and Postdocs to take on new challenges early in their careers, and we are happy to support strong candidates in applying for relevant competitive fellowships.
- 2025-2028, PI of Carlsberg Foundation Semper Ardens Accelerate grant
- 2022-2025, PI of Danmarks Frie Forskningsfond (DFF) Research Project 1 (FNU)
- 2020-2023, director of Marie Skłodowska Curie COFUND/KU TALENT PhD program
- 2020-2022, PI of Novo Nordisk Foundation, Project Grant
- 2016-2018, PI of Marie Skłodowska Curie individual grant
- 2013-2016, PI of EMBO Long-Term Fellowship
Group members
| Name | Title | Phone | |
|---|---|---|---|
| Liu, Yueyue | PhD Student | ||
| ZHANG, YONG | Associate Professor | +4552689118 |
Alumni
| Name | Title | ||
|---|---|---|---|
| Paulina Długosz | Research assistant | ||
| Muriel Leandra Schicketanz | PhD student | ||
| Cong Jiang | PhD (exchange) student | ||
| Jun Liu | PhD (exchange) student | ||
| Luokai Wang | PhD (exchange) student | ||
| Laura Juhl Jensen | Bachelor student | ||
| Yi Zhou (周翌) | MSc student | ||
| Selma Kofoed Bendtsen | Project student (Biochemistry) | ||
| Hong Jiang | Visiting researcher (MSc) | ||
| Helena Gram | Bachelor student |

