Sequencing
We have more than 10 years of experience using diverse sequencing technologies to answer microbial ecology questions.  We have developed an expertise in microbial communities analyses from a large variety of complex environmental matrices including soil, sediments and water samples but also diverse human body sites (bronchoalveolar fluids, biopsies, feces, cotton swabs). Our sequencing platform include Illumina MiSeq and MinION Oxford Nanopore.
We have developed an expertise in microbial communities analyses from a large variety of complex environmental matrices including soil, sediments and water samples but also diverse human body sites (bronchoalveolar fluids, biopsies, feces, cotton swabs). Our sequencing platform include Illumina MiSeq and MinION Oxford Nanopore.
High-throughput short-reads sequencing
Our local sequencing platform consists in a MiSeq sequencer that is mainly used for microbial communities profiling using marker genes amplicon sequencing (i.e. 16S rRNA gene, ITS2 region); small microbial draft genomes (whole genome shotgun sequencing); evaluate the functional potential of ecosystems (i.e. metagenomic shotgun sequencing) or evaluate genes expression in complex communities (i.e. metatranscriptomic).
For technical details, please visit Illumina website.
Long-reads sequencing
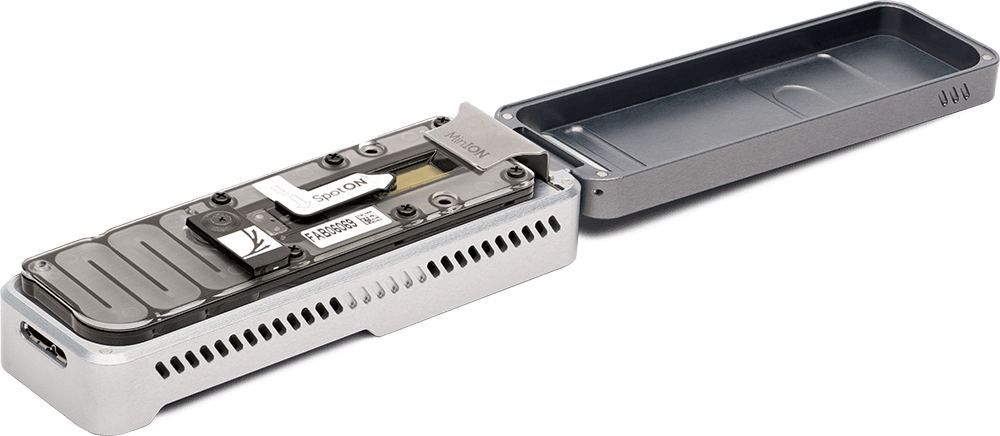 We recently acquired a MinION Oxford Nanopore sequencer. We use long-reads technology to obtain closed microbial genomes for comparative genomics application and complete plasmids molecules sequences, notably difficult to assemble using short-reads only.
We recently acquired a MinION Oxford Nanopore sequencer. We use long-reads technology to obtain closed microbial genomes for comparative genomics application and complete plasmids molecules sequences, notably difficult to assemble using short-reads only.
Section of Microbiology

Contact
Carlotta Pietroni
+45 35 33 48 41
carlotta.pietroni@bio.ku.dk
