Bacterial Cell Cycle Group - Anders Løbner-Olesen Group
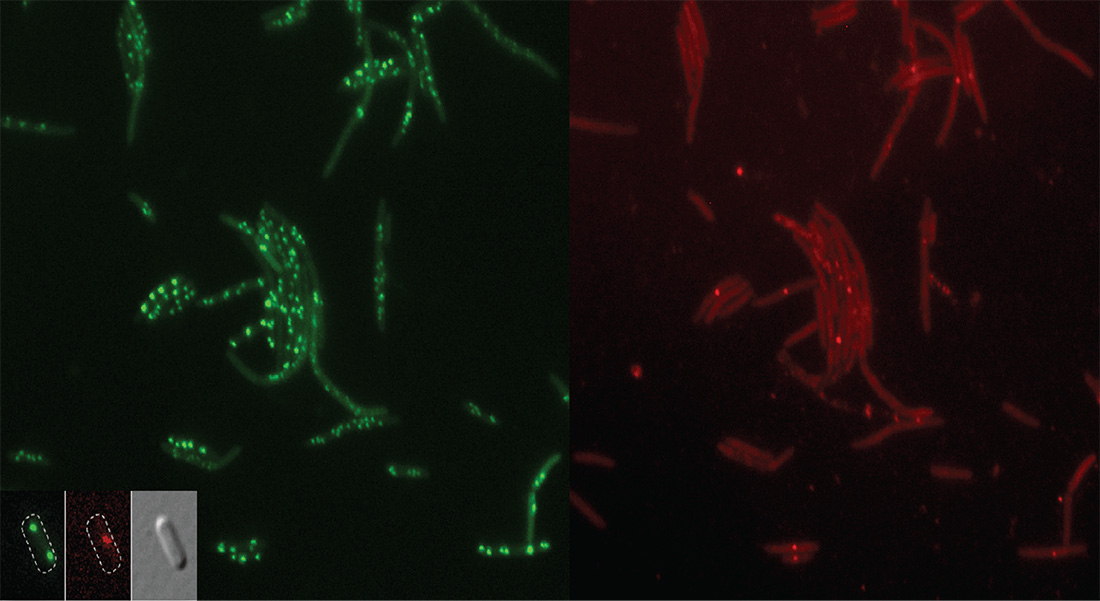
The group has historically worked on cell cycle control of DNA replication in bacteria, with focus on Escherichia coli. This remains one of the focus areas of the lab.
We have described key aspects of the E. coli cell cycle control including the DnaA protein being limiting for replication, the role of Dam, and the importance of cis-acting control regions. Recently we have widened our focus to include the development of antimicrobials with special emphasis on antimicrobial peptides.
1a. Control of bacterial chromosome replication
In most bacteria, chromosome replication is initiated by DnaA. In Escherichia coli, the frequency of initiation is controlled by the availability of active initiator protein, DnaAATP. Recent data from our lab suggests an intimate coupling between both DNA precursor biosynthesis and energy metabolism and cell cycle control.
1b. Chromosome replication during energy starvation
During steady state Escherichia coli growth, the amount and activity of the initiator protein, DnaA, control chromosome replication tightly so that initiation only takes place once per origin in each cell cycle and regardless of growth conditions. However, little is known about the mechanisms involved during transitions from one environmental condition to another or during starvation stress. ATP depletion is one of the consequences of long term carbon starvation. We have found that DnaA is cleaved in ATP depleted cells. A checkpoint at initiation of replication is apparent in such cells as no new rounds of DNA replication are initiated while already started replication events proceed to completion
2. Development of antimicrobial compounds
In recent years, many bacterial pathogens have become resistant (or insensitive) to most of the current commercial antibiotics available. We have isolated peptides that inhibit growth of both Gram-positive and Gram-negative bacteria as partners of The Danish Centre for Antibiotic Research and Development (http://dancardproject.dk/), University of Copenhagen Research Centre for Control of Antibiotic Resistance (UC-CARE) and most recently as co-founders of the Center for Peptide-Based Antibiotics (Cepan; see “Current Grants”), funded by the Novo Nordisk Foundation Challenge Programme. In CEPAN we investigate the antisense antibacterial concept for potential antibacterial compounds targeting especially the DNA replication machinery in Gram-negative bacteria as well as elucidating the mechanism of action of leading novel compounds.
Please visit “Projects” for an orientation of ongoing research project in the group.
Antimicrobial Drug Discovery and Development
Vinoth Wigneswaran
We are interested in the discovery of new antimicrobials. Specifically we are involved in the development of novel antimicrobial peptides targeting highly drug-resistant Gram-negative bacteria (E. coli, K. pneumonia, A. baumannii and P. aeruginosa). We have previously described a new antimicrobial peptide in our lab that shows promising antimicrobial activity and we are currently in the process of characterizing this compound. This includes description of mechanism of action, analysis of antimicrobial effect on synthesis of macromolecules (peptidoglycan, protein, DNA, RNA), resistance development and effect of cellular physiology. This also includes routine testing of minimum inhibitory concentration (MIC), minimum bactericidal concentration (MBC) and combinatorial testing for collateral sensitivity through synergy of antimicrobials. Furthermore, we are interested in the mechanisms involved in resistance development towards antimicrobial peptides such as colistin. These resistance mechanisms usually involve modification to the bacterial membrane, such as the addition of phosphoethanolamine and aminoarabinose to the lipopolysaccharide (LPS), which changes the membrane properties. We apply genetic manipulation, genome engineering and mass spectrometry to elucidate how these changes influence antibacterial efficacy and how these alterations may lead to collateral sensitivity, new target discovery and treatment possibilities.
Engineering biological memory in bacteria
Godefroid Charbon
The goal is to develop a new tool for information storage in bacteria using DNA methylation as an epigenetic switch. We are developing an artificial gene promoter regulated by methylation to create specific DNA methylation profiles. The novel DNA methylation patterns generated will be used to memorize inputs in living bacteria. Here we wish to design a DNA methylation system to track and record DNA replication to identify and study metabolically dormant bacteria. This system will be usable for studying bacterial phenotypic heterogeneity.
Novel antimicrobial peptide discovery
Anna Elisabeth Ebbensgaard
In response to the increasing worldwide threat to human health posed by the emergence of bacterial resistance to currently used antibiotics, the overall aim of the recently established “Center for Peptide Antibiotics” (CEPAN) is to establish a discovery platform for peptide-based antibiotics against bacterial pathogens. In particular, we are interested in the discovery of new antimicrobial peptides and identification of novel targets for peptide and PNA (peptide nucleic acid) based peptides, as well as in the discovery of bacterial envelope permeabilizing peptides (EPPs). We use intracellular peptide display to identify novel hit peptides as well as novel targets for antibiotics. Coupling intracellular peptide display to either deep sequencing or more traditional bioactivity assays, we aim to identify novel bioactive peptides targeting bacterial pathogens.
Frimodt-Møller, J., Campion, C., Nielsen,P.E., and Løbner-Olesen, A. 2021. Translocation of non-lytic antimicrobial peptides and bacteria penetrating peptides across the inner membrane of the bacterial envelope, Current Genetics, https://doi.org/10.1007/s00294-021-01217-9
Campion, C., Charbon, G., Thosen, T.T., Nielsen, P.E., and Løbner-Olesen, A. 2021. Antisense inhibition of the Escherichia coli NrdAB aerobic ribonucleotide reductase is bactericidal due to induction of DNA strand breaks. J. Antimicrob. Chemother., doi:10.1093/jac/dkab305
Frimodt-Møller, J., Boesen, T.O., Charbon, G., and Løbner-Olesen, A. 2021. Bacterial chromosomes and their replication. Molecular Medical Microbiology 3rd Ed. Edited by: Tang, Y.-W., Zhang, J.-R., Liu, D., Sails, A.D., Hindiyeh, M., and Spearman, P. Academic Press, Elsevier, Chapter 14, In press
Charbon, G., Frimodt‑Møller, J., and Løbner‑Olesen, A. 2021. Arresting coli chromosome replication upon energy starvation in Escherichia, Current Genetics, ttps://doi.org/10.1007/s00294-021-01202-2
Pereira, W., Pereira, C., Assunção, R., da Silva, I., Rego, F., Alves, L., Santos, J., Nogueira, F., Zagmignan, A., Thomsen, T., Løbner-Olesen, A., Krogfelt, K., da Silva, L., and Abreu, A. 2021. New Insights into the Antimicrobial Action of Cinnamaldehyde towards Escherichia coli and Its Effects on Intestinal Colonization of Mice. Biomolecules 2021, 11, 302, https://doi.org/10.3390/biom11020302.
Charbon, G.S., Chamizo, B.M., Campion, C., Li, X., Jensen, P.R., Frimodt-Møller, J., and Løbner-Olesen, A. 2021. Energy Starvation Induces a Cell Cycle Arrest in Escherichia coli by Triggering Degradation of the DnaA Initiator Protein. Frontiers in Molecular Biosciences, 8, 629953, doi: 10.3389/fmolb.2021.6299538
Ebbensgaard, A., Løbner-Olesen, A., and Frimodt-Møller, J. 2020. The role of efflux pumps in the transition from low-level to clinical antibiotic resistance. Antibiotics, 9, 855. doi: 10.3390/antibiotics9120855
Frimodt-Møller, J., Koulouktsis, A., Charbon, G.S., Otterlei, M., Nielsen, P.E. and Løbner-Olesen, A. 2021. Activation of the Cpx-envelope stress response system promotes tolerance to antibacterials delivered by arginine-rich peptides and aminoglycosides in Escherichia coli. Molecular Therapy: Nucleic Acid, doi: https://doi.org/10.1016/j.omtn.2021.06.009
Sinha, A.K., Løbner-Olesen, A. and Riber, L. 2020. Bacterial Chromosome Replication and DNA Repair During the Stringent Response. Front. Microbiol., 11, 582113. doi: 10.3389/fmicb.2020.582113
Jensen, S.K., Thomsen, T.T., Oddo, A., Franzyk, H., Løbner-Olesen, A. and Hansen, P.R. 2020. Novel Cyclic Lipopeptide Antibiotics: Effects of Acyl Chain Length and Position. Int. J. Mol. Sci. 21, E5829. doi: 10.3390/ijms21165829. PMID: 32823798
Center for Peptide-Based Antibiotics: Peptide Antibiotics against Resistant Bacterial Infections
Agency: Novo Nordisk Challenge program
Main applicant: Prof. Peter E. Nielsen, University of Copenhagen, Denmark
Co-applicant: Anders Løbner-Olesens (shared)
Expiration date: 2022
Link: https://cepan.ku.dk/
IRPD: A Novel in vivo Peptide Display Technology in Bacteria
Agency: Lundbeck Experiment Programme
Main applicant: Anna Ebbensgaard
Expiration date: 2023
Engineering biological memory in bacteria
Agency: VILLUM Experiment Programme
Main applicant: Anders Løbner-Olesen (shared)
Expiration date: 2022
Insane in the membrane; how to avoid crowding of the Eschericia coli inner membrane?
Agency: VILLUM Experiment Programme
Main applicant: Jakob Frimodt-Møller
Expiration date: 2022
ALO lab is involved in several different courses (both at bachelor-, master-, and Ph.D.-level) offered at the University of Copenhagen, Department of Biology, both as invited teachers but also as courses coordinators. More course information can be obtained at kurser.ku.dk.
Molecular Cell Biology (KU course code: NBIA07002U)
ALO lab share: Invited teacher (in charge of laboratory exercises).
Content: The main goal of the course is for the student to develop an understanding of how knowledge in molecular cell biology is obtained through experimental approaches; an aim particularly important and appropriate close to the point of the bachelor project.
Level: Bachelor
Credit: 15 ECTS points
General Molecular Biology (Molbiol) (KU course code: NBIA04033U)
ALO lab share: Invited teacher (in charge of laboratory exercises).
Content: The course aims at presenting the students with classical as well as modern molecular genetic terms and approaches. This course is a necessary for further enrollment in experiential-biological courses at the University of Copenhagen.
Level: Bachelor
Credit: 7.5 ECTS points
Biochemistry 1 (KU course code: LKEB10077U)
ALO lab share: Invited teacher.
Content: The course aims at presenting the students with the main topics within biochemistry, with emphasis on metabolism as well as the structure and function of important macro molecules.
Level: Bachelor
Credit: 7 ECTS points
Medical Bacteriology (KU course code: NBIK14035U)
ALO lab share: Course coordinator.
Content: To give students a thorough understanding of the field of Medical Bacteriology with emphasis on the specific traits that enables certain bacteria to cause disease. The course will cover specific bacteria, the diseases they cause and possible treatments. The course will rely heavily on genetics and molecular biology.
Level: Master
Credit: 7.5 ECTS points
Principal Subject in Molecular Genetics (KU course code: NBIA09016U)
ALO lab share: Invited teacher.
Content: The principal subject in molecular genetics deals with research that is based on phenotype/genotype relations in model organism and in humans, e.g. mutations that confer disease. Recent topics have been genome stability, cancer development, DNA replication and transcription regulation
Level: Master
Credit: 7.5 ECTS points
For public relations or our public outreach program, please contact Jakob Frimodt-Møller
Researchers
| Name | Title | Phone | |
|---|---|---|---|
| Anders Løbner-Olesen | Professor | +4535322068 | |
| Godefroid Charbon | Associate Professor | +4535322098 |
Contact
Professor Anders Løbner-Olesen
Functional Genomics
Office: 4.2.15
Ole Maaløes Vej 5
DK-2200 Copenhagen N
Email: lobner@bio.ku.dk
Phone: +45 35 32 20 68
Mobile: +45 51 43 03 26
ORCID: 0000-0002-0344-6417
