Error-free DNA repair – Lisby group
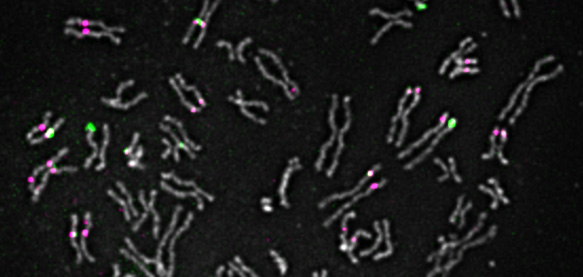
The research of my laboratory is focused on the spatio-temporal organization and regulation of homologous recombination (HR) and its role in maintaining genome integrity. We take a cell biological and genetic approach to study HR in human cells and the yeast Saccharomyces cerevisiae. Particular areas of interest are regulation of chromatin structure during HR, post-translational modification of HR proteins and the molecular architecture of HR complexes in living cells. We are particularly interested in understanding molecular mechanisms relevant to cancer predisposition, neurological disorders and premature ageing syndromes, and in utilizing homologous recombination in CRISPR-Cas9 gene-editing.
Joergensen, S., Liberti, S., Larsen, N., Lisby, M., Mankouri, H., Hickson, I. (2019) Esc2 promotes telomere stability in response to DNA replication stress, Nucleic Acids Res, 47(9):4597-4611. https://doi.org/10.1093/nar/gkz158
Bantele, S.C.S., Lisby, M., and Pfander, B. (2019) Quantitative sensing and signalling of single stranded DNA during the DNA damage response, Nature Communications, 10(1):944. https://doi.org/10.1038/s41467-019-08889-5
Crickard, J.B., Kaniecki, K., Kwon, Y., Sung, P., Lisby, M., Greene, E.C. (2018) Regulation of Hed1 and Rad54 binding during maturation of the meiosis-specific presynaptic complex. EMBO J., 37(7). https://doi.org/10.15252/embj.201798728
Pentzold, C., Shah, S., Hansen, N.R., Tallec, B., Seguin-Orlando, A., Debatisse, M., Lisby, M., Oestergaard, V. (2018) FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells. Nucleic Acids Res., 46(3):1280-1294. https://doi.org/10.1093/nar/gkx1260
Quevedo, O. and Lisby, M. (2018) Imaging of DNA ultrafine bridges in budding yeast. Methods Mol. Biol., Genome Instability, 483-493. https://doi.org/10.1007/978-1-4939-7306-4_32
Lafuente-Barquero, J., Luke-Glaser, S., Graf, M., Silva, S., Gómez-González, B., Lockhart, A., Lisby, M., Aguilera, A. and Luke, B. (2017) The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage, PLoS Genetics, 13(12):e1007136. https://doi.org/10.1371/journal.pgen.1007136
Sebesta, M., Urulangodi, M., Stefanovie, B., Szakal, B., Pacesa, M., Lisby, M., Branzei, D., Krejci, L. (2017) Esc2 promotes Mus81 complex-activity via its SUMO-like and DNA binding domains, Nucleic Acids Research, 45(1):215-230. https://doi.org/10.1093/nar /gkw882
Oestergaard, V.H., Lisby, M. (2017) Transcription-replication conflicts at chromosomal fragile sites - consequences in M phase and beyond. Chromosoma, 126(2):213-222. https://doi.org/10.1007/s00412-016-0617-2
Silva, S., Altmannova, V., Luke-Glaser, S., Henriksen, P., Gallina, I., Yang, X., Choudhary, C., Luke, B., Krejci, L. and Lisby, M. (2016) Mte1 interacts with Mph1 and promotes crossover recombination and telomere maintenance, Genes & Dev, 30(6):700-17. https://doi.org/10.1101/gad.276204.115
Moudry, P., Watanabe, K., Wolanin, K.M., Bartkova, J., Wassing, I.E., Watanabe, S., Strauss, R., Troelsgaard Pedersen, R., Oestergaard, V.H., Lisby, M., Andújar-Sánchez, M., Maya-Mendoza, A., Esashi, F., Lukas, J., Bartek, J. (2016) TOPBP1 regulates RAD51 phosphorylation and chromatin loading and determines PARP inhibitor sensitivity, Journal of Cell Biol., 212(3):281-8. https://doi.org/10.1083/jcb.201507042
The Oestergaard and Lisby groups share lab space and equipment as well as technical and theoretical knowhow. We also have joint lab meetings and joint social events. Lisby/Oestergaard group
Carl-Johan Risom gbn297@alumni.ku.dk
Oline Mathilde Pade Jensen grn726@alumni.ku.dk
Thorbjørn Nielsen kvn756@alumni.ku.dk
Henriette Iversen kcw520@alumni.ku.dk
Mads Baltser Jacobsen jcl655@alumni.ku.dk
Katrine Lundgaard jxb222@alumni.ku.dk
Simon Barlebo Wenneberg jvw478@alumni.ku.dk
Genome integrity maintenance by liquid-liquid phase separation and oscillations
Carlsberg Foundation Semper Ardens Advance grant
What?
The fundamental unit of life is the cell. We will study how cells use liquid-liquid phase separation to organize the biochemical processes of life through formation of biomolecular droplets that concentrate the enzymes and their substrates of each specific process into a confined membrane-less structure.
Why?
It remains largely unknown how liquid-liquid phase separation is achieved and regulated to ensure colocalization of enzymes with their respective substrates in specific biomolecular droplets. We expect that the outcome of this project will fundamentally change our understanding of the complexity of the cell and perhaps even the origin of life.
How?
With the proposed project, we will perform a molecular genetic dissection of liquid-liquid phase separation using DNA repair processes in the cell nucleus as a model to establish a fundamental biological understanding of how biomolecular droplets allow the cell to orchestrate its biochemical processes. We will achieve this through a cross-disciplinary effort that combines state-of-the-art microscopy with biophysical modelling and computational simulations.
Investigation of ZGRF1 as a novel anti-cancer therapeutic target
The Danish Council for Independent Research | Medical Sciences
The aim of the proposed project is to characterize the putative novel DNA repair helicase ZGRF1 and to investigate the potential of exploiting ZGRF1 in chemical synthetic lethality strategies for anti-cancer treatment. Based on our recent study of the yeast Mte1-Mph1 complex, which is the proposed homolog of human ZGRF1, we will test the hypothesis that human ZGRF1 promotes DNA repair by remodeling of homologous recombination intermediates and/or by regression/stabilization of stalled replication forks. This will be achieved by a combination of genetic analysis in human cell lines, live cell fluorescence microscopy, biochemistry and proteomics.
The role of intranuclear protein quality control in DNA replication stress recovery
The Danish Council for Independent Research | Natural Sciences
The proposed project aims to perform a molecular genetic and cell biological dissection of the role of intranuclear protein quality control in regulating Mrc1 and Pph3 during DNA replication stress recovery using yeast Saccharomyces cerevisiae as a model system. The project combines genetics, biochemistry and fluorescence live cell microscopy to dissect the molecular mechanisms that allow DNA replication restart and cell cycle progression after genotoxic stress. The evolutionary conservation of key findings will be tested using gene editing and live cell imaging of human primary cells. The importance of intranuclear protein quality control for maintaining genome integrity will be examined by measuring mutation rates, genome-rearrangements and cell survival. The results of the project will provide the framework for understanding the role of intranuclear protein quality control in the recovery from other types of stress.
Functional characterization of the ZGRF1 DNA repair helicase and its potential as a therapeutic target
The Novo Nordisk Foundation
We have discovered a new human helicase ZGRF1, which is important for repair of DNA lesions that block DNA replication. The aim of this project is to extend our genetic, cell biological and biochemical characterization of ZGRF1 and develop a ZGRF1 mouse model to examine the in vivo functions of ZGRF1. Taken together, these analyses will uncover the function and regulation of ZGRF1 and its implications for human health and disease to uncover its potential as a therapeutic target.
Carlsbergfondet (CF22-0494)
Novo Nordisk Foundation (NNF19OC0055203)
Independent research fund Denmark (7016-00214)
Independent research fund Denmark (7014-00176)
Villum Foundation (11407)
ERC starting grant (242905)
Staff
| Name | Title | Phone | |
|---|---|---|---|
| Hoa Phan | Laboratory Technician | +4535331872 | |
| Michael Lisby | Professor | +4535322120 |
Contact

Principal investigator
Professor Michael Lisby
Section for Functional Genomics
Ole Maaloees Vej 5
DK-2200 Copenhagen N, Denmark
Email: mlisby@bio.ku.dk
Phone: +45 3532 2120
