Welcome to the Sine Lo Svenningsen Group
Our group has close ties to the rest of the Biomolecular regulation group. We are particularly interested in exploring two aspects of bacterial survival strategies, namely the rapid transcriptional and translational reprogramming that occurs in E. coli in response to starvation, and how phenotypic heterogeneity within a population of stressed cells may influence their survival.
Contact Sine if you are interested in our work – we always welcome applications to join the lab. You can read more about current research projects below and read more about joining the lab as a student on the Section for Biomolecular Sciences student page.
Our research interests centers on bacterial gene regulation. The components and interaction partners of gene regulatory networks are being identified at a breakneck pace in genome-wide studies in many model organisms. In the post-genomic era, a central challenge is to understand the processes that govern the dynamics of such networks. We are interested in all aspects of gene regulatory network analysis, from dissecting detailed molecular mechanisms of regulation to determining the logic and dynamic behavior of the network as a whole. In the group, we study either synthetic networks, or existing networks that control behaviors where timing and network dynamics are expected to be particularly critical. This is true, for example, of the networks regulating bacterial cell-cell communication (quorum sensing), bacterial stress responses, or bacteriophage developmental decisions. On most projects, we collaborate with biophysicists to develop theoretical models that help us answer fundamental physiological or mechanistic questions in a quantitative manner.
In the Svenningsen group, we combine techniques from molecular biology, genetics, growth physiology and mathematical modelling to address fundamental questions in gene regulation, especially concerning RNA-centered regulatory mechanisms. We strive to always put the study of detailed molecular mechanisms into the context of the behavior of the bacterial population as a whole. The main research projects are:
Dynamic regulation of the tRNA pool
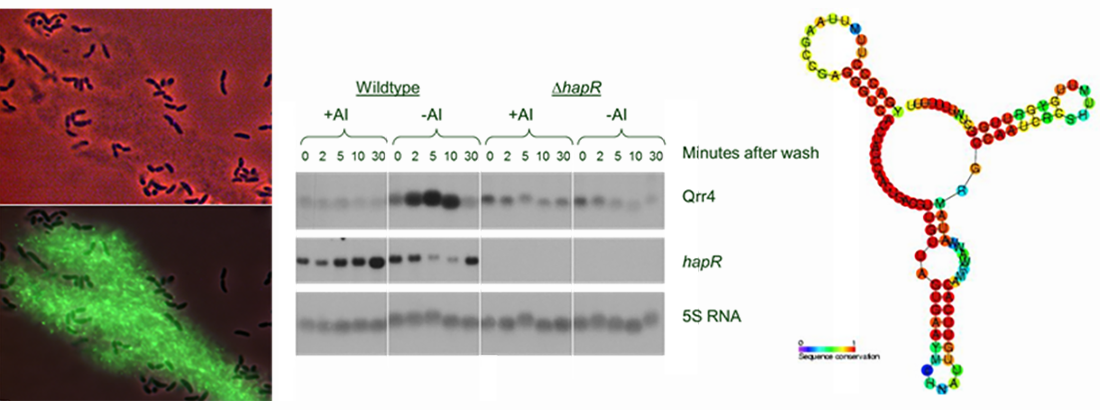
Transfer RNAs (tRNA) are the most numerous noncoding RNAs found in the cell, and serve a key role in protein synthesis. Using an improved method of quantification, we have recently demonstrated highly dynamic regulation of tRNA levels in the model bacterium Escherichia coli. It turns out that this bacterium is continually adjusting the levels of available tRNA to match the current demand for tRNA in protein synthesis, by breaking down excess tRNA. Our focus is now on understanding how this demand-based regulation of tRNA levels occur, and how it helps bacteria cope with poor growth conditions.
Quorum sensing control of phage-bacterial interactions
Bacteria have developed a multitude of mechanisms to protect themselves from attack by phages. We find that E. coli upregulates an anti-phage defense mechanism when growing in crowded environments, as indicated by the presence of quorum-sensing signaling molecules produced by surrounding bacteria. A more sophisticated example of this strategy was found in the fish pathogen Vibrio anguillarum, which chooses between different anti-phage defense mechanisms dependent on quorum-sensing signals. We are currently focused on a marine prophage that appears to eavesdrop on the host cell’s quorum sensing system to goven its decision to induce lytic development. We are also interested in how the spatial structures that are developed as bacteria grow (e.g., microcolonies, biofilm) impact phage-bacterial interactions, and have recently shown that microcolonies can sustain growth while undergoing attack by a lytic phage, because the growth of bacteria throughout the spherical colony exceeds the killing by phage on the surface.
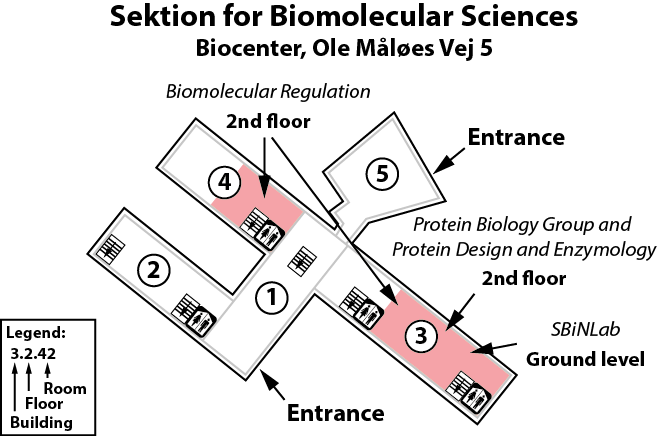
The lab is located on the 2nd floor, building 3 and 4 of the Copenhagen Biocenter (map), close to the centre of Copenhagen. It takes less than 1 hour to travel from Copenhagen's airport to our lab, by train or subway (from the airport to Nørreport station, a 20 minute journey, with trains every 10 minutes) and bus (bus 150S from Nørreport to the stop called "Fredrik Bajers Plads" in the intersection of Tagensvej and Nørre Allé, a 10 minute journey, with buses every 6-8 minutes). From there it is a few minutes walk.
Contact
Associate professor Sine Lo Svenningsen
Section for Biomolecular Sciences
Kaj Ulrik Linderstrøm-Lang Centre for Protein Science
Ole Maaløes Vej 5, room 4-2-27
DK-2200 Copenhagen N
Email: sls_at_bio.ku.dk
Phone: +45 3532 2033
KU profile: Link