Organization of Metabolism in the Hvorecny Group

We investigate the structure-function mechanisms in proteins that underlie disease and generate species-specific biological traits across the domains of life, pairing biochemical and structural studies with studies at the cellular and organismal level. To accomplish this, we apply a variety of techniques at a range of biological scales, including cryo-EM, cryo-ET, biochemical and biophysical assays, and tissue cell culture. Past work includes defining the substrate profile and in vivo effects of a bacterial virulence factor, helping to settle a long-running debate about the cytoskeletal protein actin from the parasite T. gondii, and identifying a previously unobserved antibody-binding mechanism. Current work focuses on the physical organization of metabolism. Read more below.
Interested in collaborating or joining our team? Feel free to reach out to Kelli to discuss – we always welcome ideas and applications!
Every cell rapidly responds to changing conditions by altering its flow of energy and molecular building materials to meet new demands. Cells can swiftly alter the flow of entire pathways in metabolism by triggering the reversible assembly of enzymes into micron-scale structures. These assemblies, many of which are filaments, modify the shape of constituent enzymes, tuning the rates of molecule synthesis and redirecting metabolic materials. While researchers recognize the importance of this cellular response, we lack basic principles describing how a cell organizes these assemblies and how assemblies diverge throughout evolution. We employ biophysical, biochemical, and cellular approaches to define the fundamental principles that physically organize metabolism the cell.
Current questions include:
- How do metabolic enzymes combine regulatory mechanisms (filament assembly, post-translation modifications, isoform and gene co-assembly, small molecule activation and inhibition) to modulate metabolic output?
- How do environmental signals trigger assembly and disassembly in vivo and what are the downstream effects?
- How are these regulatory principles modified across the tree of life?
The spaciotemporal principles of metabolism are required to understand and develop treatments for metabolic diseases and disorders, including rare genetic diseases, metabolic dysregulation, and cancer. Assembly differences between humans and pathogenic organisms can potentially be leveraged for new antimicrobial strategies. In addition, the fundamentals of protein assembly can be applied to various design problems, which include creating green biofactories for molecules, developing enzyme-based chemical remediation platforms, and the de novo generation of new protein assemblies.
We are always excited to receive applications from potential postdocs and students (PhD, Masters, or Bachelors)! If you are interested in joining the laboratory, reach out to Kelli to discuss position availability and specific projects.
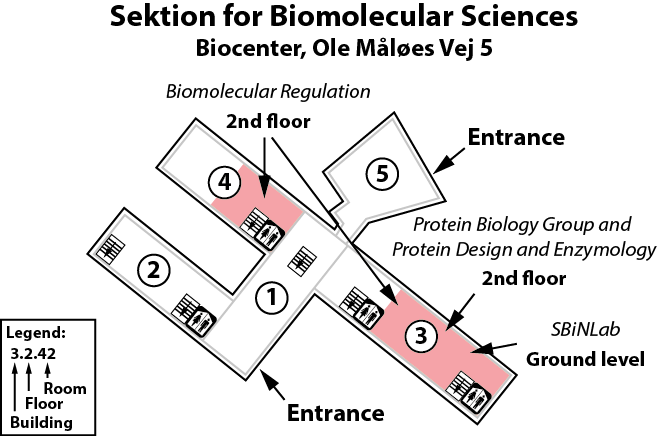
Our lab is located on the 2nd floor of building 3 of the Copenhagen Biocenter (map), close to the center of Copenhagen. It takes less than 1 hour to travel from Copenhagen's airport to our lab, by train or subway (from the airport to Nørreport station, a 20 minute journey, with trains every 10 minutes) and bus (bus 150S from Nørreport to the stop called "Fredrik Bajers Plads" in the intersection of Tagensvej and Nørre Allé, a 10 minute journey, with buses every 6-8 minutes). From there it is a short walk.
Contact

Assistant professor Kelli Hvorecny
Section for Biomolecular Sciences
Kaj Ulrik Linderstrøm-Lang Centre for Protein Science
Ole Maaløes Vej 5, room 3-2-47
DK-2200 Copenhagen N
Email: kelli.hvorecny_at_bio.ku.dk
Phone: +45 3532 1500