Welcome to the Martin Willemoës website
In the Martin Willemoës group we work with enzymology and structural biology. We have worked for many years with the molecular genetics of E. coli and other bacteria.
Within the last five years our research has been dedicated to the coupling of in silico design with molecular evolution based on selection systems for selecting enzyme activity.
My group is hosting a state of the art isothermal titration calorimetry facility allowing analysis in small volumes. We are using it to study the kinetics of enzyme catalyzed reactions, protein-protein interactions and protein-ligand interactions and wellcome proposals for collaboration in this fileld.
As a student you are also very welcome to stop by my office to discuss project proposals with me. Read more about how to join the group on the Section for Biomolecular Sciences homepage on joining the labs.
We combine enzymology and enzyme design with the development of genetic selection systems based on the growth of Escherichia coli and Lactococcus lactis to evolve new enzyme activities directed towards e.g. new peptide recognition sequences (HIV protease), deglycosylation of glyco-proteins and the mechanisms that guide the specificty excerted by binding modules and -sites in reactions catalysed by maybe otherwise promiscous endo- or exoglycosidases. These projects in part serve to make us wiser with respect to the rules governing the design of enzymes that can lead to a whole new tool-box addressing the problems faced with green chemistry, industrial waste etc. One example is the design of a new enzyme improved alternative to Casein used in the danish dairy industry.
We are working on the following projects:
- Specificity and drug resistance of virus proteases
- Basic characterization of enzymes
- High-throughput screening
- Collaborative projects
Specificity and drug resistance of virus proteases
We also try to gain insight into e.g. the complex rules that govern substrate specificity or development of resistance to anti-HIV drugs in HIV protease. Likewise, we also work on the M2 proton transport channel of the influenza virus to elucidate the mechanisms by which resistance to drugs by mutation also takes place for this channel. Hopefully, we may be able to predict the efficiency of drugs from the combination of data and computational chemistry. These projects take place in a close collaboration with Assoc. Prof. Kresten Lindorff-Larsen and Prof. Jakob R. Winther.
Basic characterization of enzymes
Many of the projects also involve basic characterisation of enzymatic properties. An example could be alpha-amylase I from barley, where we study the complex kinetics of amylopectin degradation. Enzymes from the nucleotide metabolism that we study involve the orotate phosphoribosyltransferase where we study in great detail the catalytic mechanism to an almost nerdy degree of details and also the bifunctional dCTP deaminase:dUTPase from M. tuberculosis is an example of an enzyme for which the subtle and intriguing mechanism of regulation has produced several projects over the years.
High-throughput screening
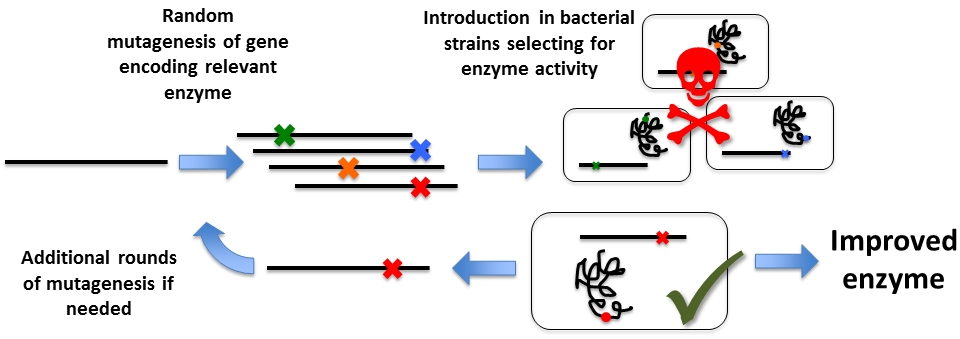
A third area of interest is to figure out good methods of generating mutant libraries with as many different mutations represented as possible. We have designed a system in which we perform error-prone PCR on the beta-lactamase gene (the enzyme that confers ampicillin resistance) to generate large libraries from which to select for mutations that promote the ability of the mutant enzyme to break down other antibiotics of the ampicillin family. Mutations and associated enzyme activities that are also known from multi-resistant pathogenic bacteria. At the same time, because beta-lactamase represents a good selection system, we work to unravel by multifactorial analysis the mechanisms guiding the outcome of mutations from varying the reaction conditions for error-prone PCR. This is done in order to generate a general description and effective protocol for error-prone PCR, regardless of template properties.
Collaborative projects
Recently, we have initiated a collaboration with collegues in our the section and headed by Prof. Karen Skriver on intrinsically disordered proteins, IDP's, that are involved in transcriptional regulation in plants. The project addresses the interesting question of how a protein can transform into a highly specific signal mediator through interaction with other protein partners, despite a seeming lack of structure. We contribute to the collaboration mainly via our expertise in microcalorimetry, where we investigate and assist in interpretating the thermodynamic parameters determined for the IDP's and their interaction with binding partners. Projects within this subject will typically be co-supervised with Karen Skriver who is the expert on the particular proteins and their related biological context.
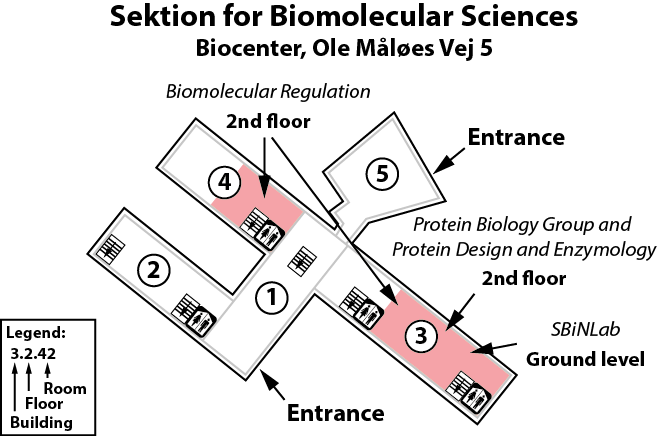
Our lab is located on the 2nd floor, building 3 of the Copenhagen Biocenter (map), close to the centre of Copenhagen. It takes less than 1 hour to travel from Copenhagen's airport to our lab, by train or subway (from the airport to Nørreport station, a 20 minute journey, with trains every 10 minutes) and bus (bus 150S from Nørreport to the stop called "Fredrik Bajers Plads" in the intersection of Tagensvej and Nørre Allé, a 10 minute journey, with buses every 6-8 minutes). From there it is a few minutes walk.
Contact

Associate professor Martin Willemoës
Section for Biomolecular Sciences
Kaj Ulrik Linderstrøm-Lang Centre for Protein Science
Ole Maaløes Vej 5, room 3-2-47
DK-2200 Copenhagen N
Email: willemoes_at_bio.ku.dk
Phone: +45 3532 2030
KU profile: Link