Welcome to the Lindorff-Larsen Group
My main focus is protein dynamics and their relationship to protein structure and function. We study these by integrating computational methods with experimental studies, to understand how the ability of proteins to change their shape help modulate, or indeed determines, their function. Read more about my research below.
My group is highly international and cross-disciplinary with most students having an additional supervisor with an eperimental background. Thus, my group is part of both the Protein Biology Group and the Structural Biology and NMR Laboratory.
If you are interested in establishing a collaboration with my group or do a project in my group, feel free to stop by or contact me by mail or phone.
My research deals broadly with the relationship between protein structure, dynamics and function. We mostly use computational methods, generally integrated very tightly with experimental studies, to understand how the ability of proteins to change their shape help modulate, or indeed determines, their function.
We have contributed to methods for integrating NMR spectroscopy and atomistic simulations to study the motions of both folded and intrinsically disordered proteins and more recently RNA. We have demonstrated how simulations can be used to elucidate slow motions in proteins and capture the mechanism, thermodynamics and kinetics of conformational change and ligand binding processes. We were also the first to determine the structures of fibrillar proteins using sequence coevolution and NMR.
Within protein folding we have pushed the boundaries of how simulations can be used to study folding mechanisms. We have developed simulation methods and some of the most accurate force fields for simulations, that are now used by thousands of researchers worldwide. Together, these developments enabled us to provide unprecedented mechanistic insight and detail of the process of protein folding. More recently, we have shown how simpler models can be used to study the complexities of membrane protein folding , and how models of protein folding and evolution can be used to understand how diseases develop.
PRISM |
PRISM - Protein Interactions and Stability in Medicine and Genomics PRISM is a Novo Nordisk Foundation Challenge Centre on protein chemistry. The driving force behind PRISM (Protein Interactions and Stability in Medicine and Genomics) is the convergence of recent developments within seemingly unrelated areas: structural biology, protein folding, high-throughput experiments, computational biophysics and genomics. The overarching hypothesis behind PRISM is that by combining these developments we may transform our current view of how proteins fold and function inside cells, how mutations perturb these processes and how we can use such information to understand, diagnose and ultimately treat human diseases. In this way, PRISM aims to move studies of protein folding and stability into a cellular context, and to use the resulting understanding to further human health. |
 |
BRAINSTRUC - Complex Structural Aspects of Signalling, Inflammation and Disease in the Brain BRAINSTRUC was a Lundbeck Foundation initiative on integrative structural biology. The partners in the BRAINSTRUC consortium were experts in complementary structural biology disciplines and spearheaded this revolution in Danish research, enabling very complex biological questions to be answered. One of the most challenging questions concerned the dynamic interplay between proteins, membranes and other macromolecules in the brain. It was enabled by the new ground-breaking facilities, BRAINSTRUC investigated the structural and functional facets of 1) the innate immune system in the brain and its role in autoimmune neurological disease; 2) the role of ion-pumping and lipid-flipping proteins in neuronal signalling and overall structure of nerve cell membranes; 3) the mechanism of protein fibril formation in Parkinson’s disease and their correlation with pathogenesis. |
Being of SBiNLab we have access to state-of-the-art equipment for protein production, purification and characterization. In addition, our research requires substantial computational resources, and through several grants we have been able to purchase >1000 computing cores that we have access to locally. In addition, we have access to the Danish national high-performance computing facility Computerome 2 and have, as one of the only Danish biocomputing groups, been awarded computing time (twice) via the competitive European “Partnership for advanced computing in Europe (PRACE)”. At all these sites we have access to excellent technical support, in addition to the substantial experience that has been established within my group.
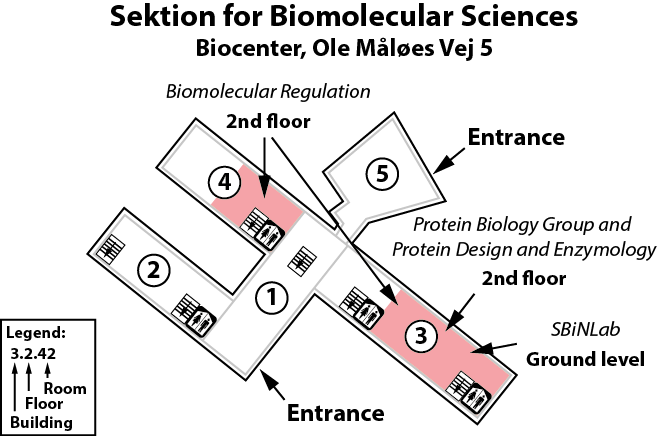
The Lindorff-Larsen lab is located in the Copenhagen Biocenter (map) next to the main entrance of building 1 at the ground floor and in the middle of building 3 on the second floor. The BioCenter is located close to the centre of Copenhagen. It takes less than 1 hour to travel from Copenhagen's airport to our lab, by train or subway (from the airport to Nørreport station, a 20 minute journey, with trains every 10 minutes) and bus (bus 150S from Nørreport to the stop called "Fredrik Bajers Plads" in the intersection of Tagensvej and Nørre Allé, a 10 minute journey, with buses every 6-8 minutes). From there it is a few minutes walk.
Contact

Professor Kresten Lindorff-Larsen
Section for Biomolecular Sciences
The Kaj Ulrik Linderstrøm-Lang Centre for Protein Science
Structural Biology and NMR laboratory
Ole Maaløes Vej 5, room 3-2-11
DK-2200 Copenhagen N
Email: lindorff_at_bio.ku.dk
Phone: +45 3532 2027
KU profile: Link