
REPIN is a cross-disciplinary endeavour with the ambition to widen the definitions and rules for protein interactions. This project aims to foster a lasting renaissance in basic and applied protein chemistry encompassing intrinsically disordered proteins.
Research and discoveries are mostly driven from established concepts and searches around normality. Together with mental borders from paradigms, this causes rare advances and breakthroughs. The lock-and-key model for protein interactions postulated in 18941 followed by the induced-fit model in 19582 have been instrumental for our understanding of molecular communication and effective for drug developments. However, with the discovery of intrinsically disordered proteins (IDPs), these models have met limitations and research into how molecular interactions can be achieved in the absence of form is critically required.
NEWS:
- Postdoc in Mechanisms of Signalling and Transcription by Structural Disorder (application deadline: 10th March 2023)
- NEW VIDEO ABOUT IDP RESEARCH FROM REPIN OUT NOW: LINK
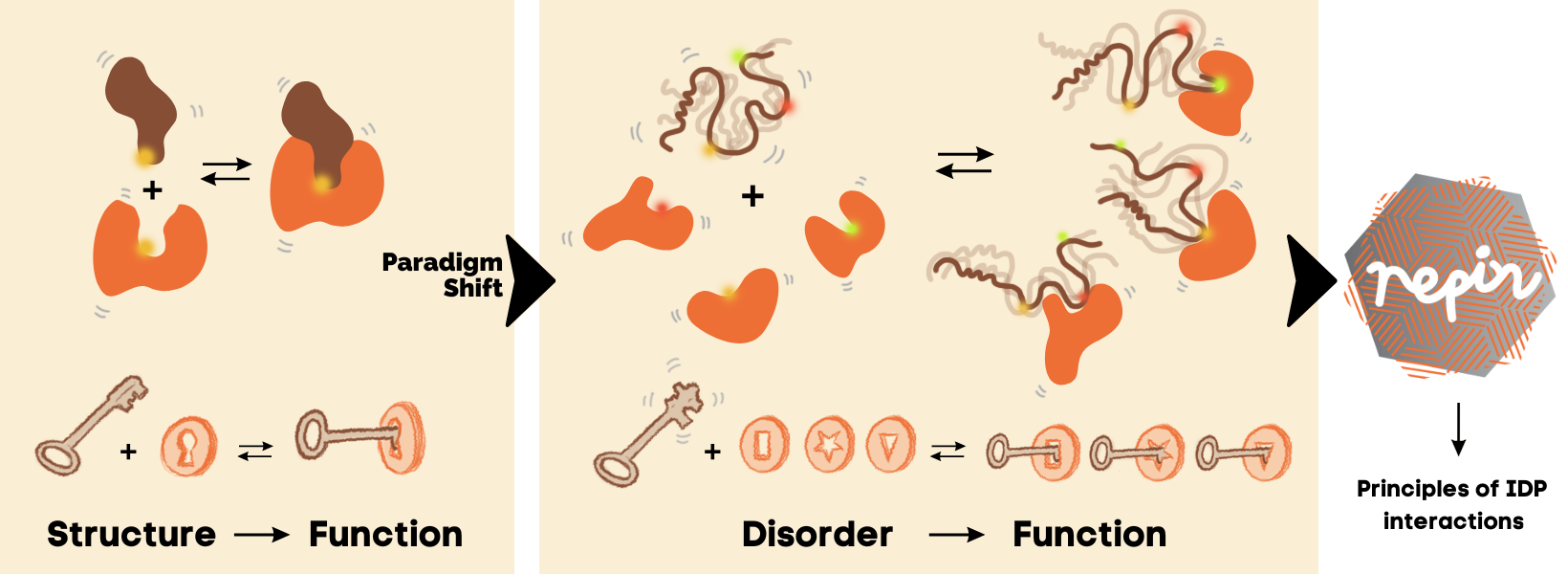
In REPIN, we take on the challenge to uncover hitherto unknown principles of intrinsic disorder (ID)-based protein interactions. We do this by developing novel concepts and by fostering a renaissance of classic protein chemistry terms including specificity, allostery, polyelectrolyte behaviour, and allovalency by disorder. The understanding of molecular communication can be revolutionized but it requires a rethinking of protein interactions in the frame of molecular disorder.
New concepts and vocabulary in protein chemistry await discovery from studies of carefully selected ID-based interaction model systems.
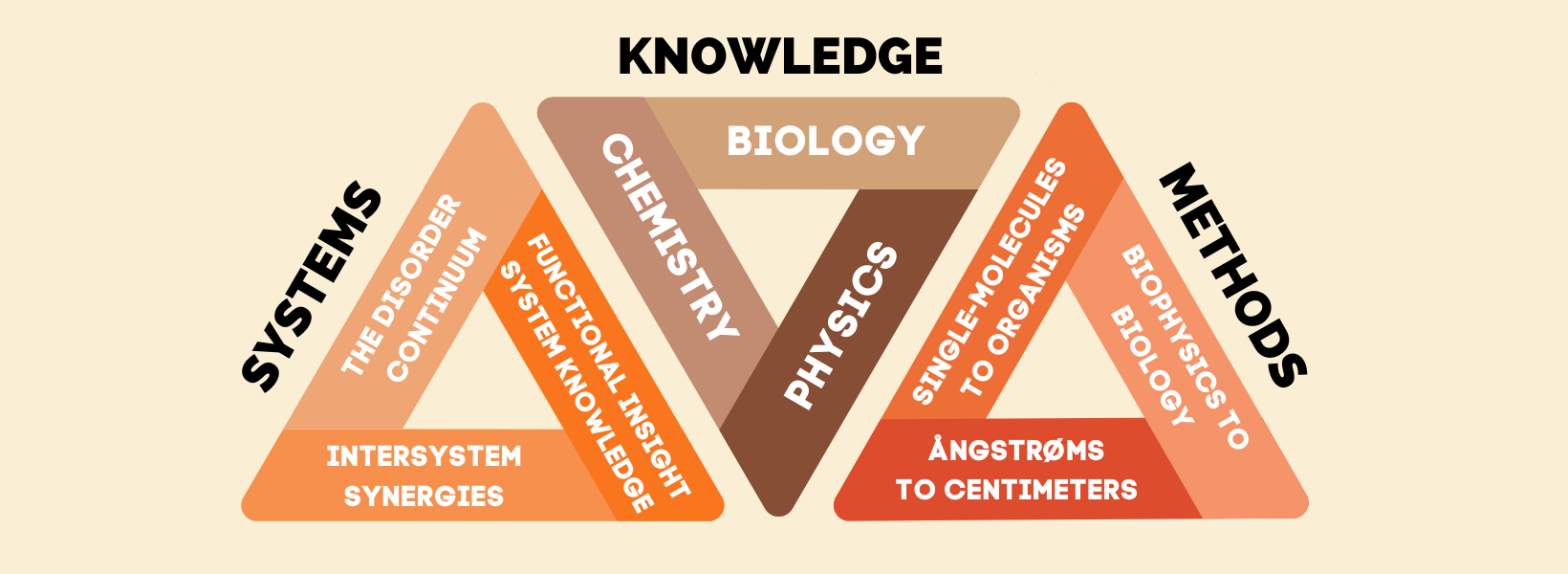
THE IDEA is to establish a multi-scale experimental protein chemistry strategy ranging from studies at the single molecule level over ensembles to whole organisms, to capture features of ID-based interactions, which manifest themselves at different levels of resolution. The know-how and experimental and theoretical aptitudes in REPIN enable us to approach disorder in multiple selected systems covering all time- and length scales at depth.
The vision is to change our conception of molecular communication through overcoming contemporary challenges in protein interactions.
The aims are to develop concepts and models for ID-based interactions that incorporate new as well as classic protein chemistry terms.
Our ambitions are to widen the definitions and rules for protein interactions and to foster a lasting renaissance in basic and applied protein chemistry encompassing ID.
REPIN is funded by a Novo Nordisk Foundation Challenge grant.
 The REPIN consortium includes Prof. Birthe B. Kragelund (BBK), Prof. Benjamin Schuler (BS), Prof. Karen Skriver (KS), and Prof. Rasmus Hartmann-Petersen (RHP) and includes a core collaborative base of Prof. Lise Arleth (LA) and Robert Best (RB). Expert competences cover structural biology, protein chemistry, protein folding, biophysics, biochemistry, genetics, and applied bioinformatics.
The REPIN consortium includes Prof. Birthe B. Kragelund (BBK), Prof. Benjamin Schuler (BS), Prof. Karen Skriver (KS), and Prof. Rasmus Hartmann-Petersen (RHP) and includes a core collaborative base of Prof. Lise Arleth (LA) and Robert Best (RB). Expert competences cover structural biology, protein chemistry, protein folding, biophysics, biochemistry, genetics, and applied bioinformatics.
Birthe B. Kragelund, ORCID: 0000-0002-7454-1761, is professor of biostructural NMR spectroscopy at the University of Copenhagen and head of REPIN. From a background in protein NMR spectroscopy, protein folding and protein chemistry, and educated at the Carlsberg Laboratory, BBK has worked to further the understanding of protein disorder and functional dynamics and their impact on biology. Through cross-disciplinary collaborations, she has propagated knowledge to fields outside structural biology, and pioneered the characterization of intrinsically disordered proteins (IDPs) in particular in membrane proteins. Her research addresses protein intrinsic disorder at several levels, and includes the associated fields of protein folding and interactions, where she focuses on disorder in cellular signaling through integrative methods. Included in her 100+ peer-reviewed papers, she has also published protein chemistry methods, reviews that bridge across fields, book chapters, as well as educational outreach papers.
BENJAMIN SCHULER, ORCID: 0000-0002-5970-4251, is professor of Molecular Biophysics at the University of Zurich, CH, and has a background in biochemistry and biophysics. BS is an expert in protein structure and dynamics, especially using single-molecule spectroscopy. By developing new single-molecule methods and integrating them with a broad range of complementary biochemical and biophysical techniques, he has been able to establish a quantitative understanding of IDPs and protein conformational dynamics on
previously inaccessible length- and timescales. Recently, BS has started to focus on the functional properties of IDPs, especially their interactions with other biomolecules. Key competences include protein biophysics, single molecule spectroscopy, IDPs, protein folding and protein dynamics.
RASMUS HARTMANN-PETERSEN, ORCID: 0000-0002-4155-7791, is professor at BIO, UCPH. With a background in protein chemistry and yeast genetics he has established himself as an internationally recognized expert in regulated protein degradation. Through biochemical, structural and genetic studies RHP has identified and characterized several proteins involved in the ubiquitin-proteasome system, including new proteasome subunits and co-factors, a novel intrinsically disordered ubiquitin-binding protein, which is highly multifunctional and broadly involved in degradation and signaling. Key competences include IDPs, genetic screens and proteomics, and detailed cell biological, structural and biophysical studies.
KAREN SKRIVER, ORCID: 0000-0003-2225-4012, is professor at BIO, UCPH, and with a background in protein chemistry, biochemistry, and plant biology KS has established herself as an internationally recognized expert in macromolecular interactions within transcriptional networks. Through biochemical, structural and transgenic studies KS has characterized functional elements of gene-specific transcription factors (TFs) regulating stress responses and development. This includes identification of a DNA-binding-site landscape and regulatory network for NAC transcription, structures and linear motifs of intrinsically disordered NAC regions providing clues to the interactome of a cellular and evolutionary patterns of disorder profiles, and whole organism effects of changed TF-levels in vivo. Her studies contribute to a platform for revival of classical biochemical concepts, developed for folded proteins, in the light of ID. Key competencies include biochemical and protein chemistry approaches followed by detailed structural, biophysical, and whole organisms in vivo studies.

REPIN offers an ambitious and inspiring crossdisciplinary workplace with engaged and enthusiastic colleagues and challenging projects. If you wish to join our venture, feel free to contact one of the core members and present them with your project, or hear about openings in existing or starting projects - we always have bachelor or master's projects at hand.
Occasionaly, we also have funded PhD or postdoc positions available. In that case, you can find more information and apply on the UCPH job portal: https://employment.ku.dk/
CoIN is an association of researchers in Copenhagen with a focus on intrinsically disordered proteins (IDPs) in biology. CoIN wishes to increase the awareness towards IDPs in their own as well as in associated fields through the ambition to have open talks and workshops on the conformation properties and functional aspects of IDPs.
The concentration of researchers working on IDPs is particularly high in the greater Copenhagen area and network meetings are arranged to stimulate exchange of results and ideas and to foster new research directions and formulation of novel research questions across disciplines, faculties and institutions.
Currently the network connects research ranging from computational analyses to whole organism studies.
CoIN members
Birthe B. Kragelund, Linderstrøm-Lang Centre, University of Copenhagen
Homepage: www.bio.ku.dk/bms/bbk
IDP relevant research: The Kragelund group combines biophysical studies with cell-biology to study how intrinsic disorder underlie cellular decision making. In particular they address the role of intrinsic disorder in membrane proteins and study the interplay with membrane constituents and signaling molecules. Mainly based on NMR methodology in combination with a suite of other biophysical techniques they aim to decipher how fuzziness is deterministic for protein recognition and protein function.
Elena Papaleo, Danish Cancer Society
Homepages: https://www.cancer.dk/research/dcrc-research-units-and-groups/computational-biology-laboratory/, https://sites.google.com/site/cblcancerdk/IDP relevant research: Elena Papaleo is the group leader of the Computational Biology Laboratory (CBL) at the Danish Cancer Society (DCRC). CBL research focuses on the study of conformational ensembles of intrinsically disordered proteins or disordered tails, loop and linker of folded proteins that play crucial roles in cancer pathways. We are especially interested in the effects induced upon post-translational modifications and cancer-related mutations on the conformational ensemble of IDPs in their free and bound states. We are also studying key scaffolding proteins working at the cross-road between autophagy and cell proliferation in collaboration with the Unit of Cell Stress and Survival and the Unit of Cell Death and Metabolism at DCRC. We use enhanced sampling molecular dynamics simulations (including metadynamics, NMR-driven sampling and replica exchange) and bioinformatics tools to unravel PTM-modulated structural motifs in IDPs. We work in tight collaborations with experimental groups from different fields, including cellular biology, biochemistry, and biophysics in Denmark and abroad.
Karen Skriver, Linderstrøm-Lang Centre, University of Copenhagen
Homepage: https://www.bio.ku.dk/bms/ks/IDP relevant research: Intrinsically disordered transcription factors and folded hub proteins
Our studies focus on the mechanism of interactions between intrinsically disordered plant transcription factors and structured cellular hubs. The results from biophysical characterization are translated to the organismal level to improve understanding of essential cellular interactomes.
Homepage: https://www.bio.ku.dk/bms/kllIDP relevant research: The Lindorff-Larsen group combines molecular dynamics and Monte Carlo simulations with experiments to study the relationship between protein structure, dynamics and function. A key goal is to be able to provide a realistic and useful description of the complicated ensembles of conformations a protein may take, and to use this to understand its biological behaviour. In the context of intrinsically disordered proteins and intrinsically disordered regions we are developing novel methods to sample these efficiently by computation, and to use information from e.g. NMR and SAXS to understand how structural preferences are determined and how they affect functions such as binding and regulation.
Homepage: https://www.bio.ku.dk/staff/ellgaardIDP relevant research: We are interested in the role of IDP’s in recognizing substrates for proteasomal degradation, for instance IDP’s present in proteins of the ER-associated degradation (ERAD) pathway and in E3 ubiquitin ligases.
Martin Willemoës, Linderstrøm-Lang Centre, University of Copenhagen
Homepage: https://www.bio.ku.dk/bms/mwIDP relevant research: We use isothermal titration calorimetry to determine the thermodynamic paramters of intrinsically disordered proteins. The advantage of ITC in studying peptide and protein interactions within IDP domains is that no labeling is required and that the true thermodynamics of the interactions are obtained. This allows us to analyse the entropi and enthalpy effects of side chain substitutions and suggest mechanisms for the interaction between ID regions and their binding sites.
Meike Burow, Center of Excellence for Dynamic Molecular Interactions (DynaMo), University of Copenhagen
Homepage: https://dynamo.ku.dk/people/burowIDP relevant research: “Plant metabolism is continuously evolving new biosynthetic pathways that provide novel biotic and abiotic resistance mechanisms, but relatively little is known about the corresponding evolution of the necessary regulatory factors. We study how new and specific regulatory patterns arise from changes in the composition of transcription factor complexes and whether this occurs through rapid evolution of transiently structured protein regions.”
Homepage: https://www.cancer.dk/research/statistics-bioinformatics-registry/sbrresearch/IDP relevant research: IDP oncogenic CIP2A and PP2A involved in metabolism and autophagy regulation in cancer.
Inhibition of protein phosphatase 2A is an hallmark of cellular transformation. Cancer cells express highly IDP proteins such as CIP2A and SET to directly suppress PP2A activity engaged to control major growth, survival and cell death signaling pathways. Little is known about these endogenous highly expressed PP2A inhibitor CIP2A and SET functions. Recently, we investigated the role of CIP2A in cancer cell transformation and survival. We characterized mechanism behind CIP2A in regulating of mTORC1, c-Myc and AMPK pathways, three major cancer cell metabolism regulating pathways, trough PP2A inhibition. Currently, we characterize structural diversity and numerous post-translational modifications, such as phosphorylation, ubiquitination and acetylation of CIP2A in order to understand how it associates with such a major signaling nexuses. Also, what is the molecular regulatory and inhibitory mechanism behind association between CIP2A and PP2A. Characterization of functional motifs that serve as a platform between CIP2A, PP2A, AMPK, mTORC1 and c-Myc association is our goal.
Rasmus Hartmann-Petersen, Linderstrøm-Lang Centre, University of Copenhagen
Homepage: https://www.bio.ku.dk/bms/rhpIDP relevant research: We focus on the role of IDPs in intracellular protein degradation via the ubiquitin-proteasome system, using yeast genetics, proteomics and structural techniques to probe their function.
Stine Falsig Pedersen, Cell and Developmental Biology, University of Copenhagen
Homepage: https://www1.bio.ku.dk/staff/FalsigPedersen/IDP relevant research: The core interest of the Pedersen group is acid-base transport proteins of the solute carrier (SLC) superfamily. Our strategy is to study a few selected SLCs using a “vertical” approach in which we address questions from the regulation of their transcription and translation, their posttranslational regulation, structure, cell biology, and finally to their function and pathology-related dysfunction in animal models. This allows us to use, for instance, understanding of how posttranslational regulation affects SLC protein structure, dynamics, and interactions, to gain mechanistic insight in the dysregulation of these proteins in disease. The SLC family most extensively studied in our group is the SLC9A family of Na+/H+ exchangers, which exhibit a partially evolutionally conserved intrinsically disordered (ID) region in their C-terminal tail, which we have shown is very important for the regulation and function of central members of this family. Our current focus is to elucidate how evolution of ID in the SLC9A family may underlie the differences in localization, regulation and function of the various family members, with particular focus on how ID orchestrates molecular “hub” functions of this important family of transporters.
Funded by:
REPIN has received six year funding from the Novo Nordisk Foundation Challenge programme
Project: REPIN - Rethinking Protein-Protein Interactions
Period: 2019-2025
Contact
Prof. Birthe B. Kragelund
Copenhagen Biocenter
Ole Maaløes Vej 5
DK 2200 Copenhagen N
Phone: +4535322081
E-mail: bbk@bio.ku.dk
KU research profile: Link

